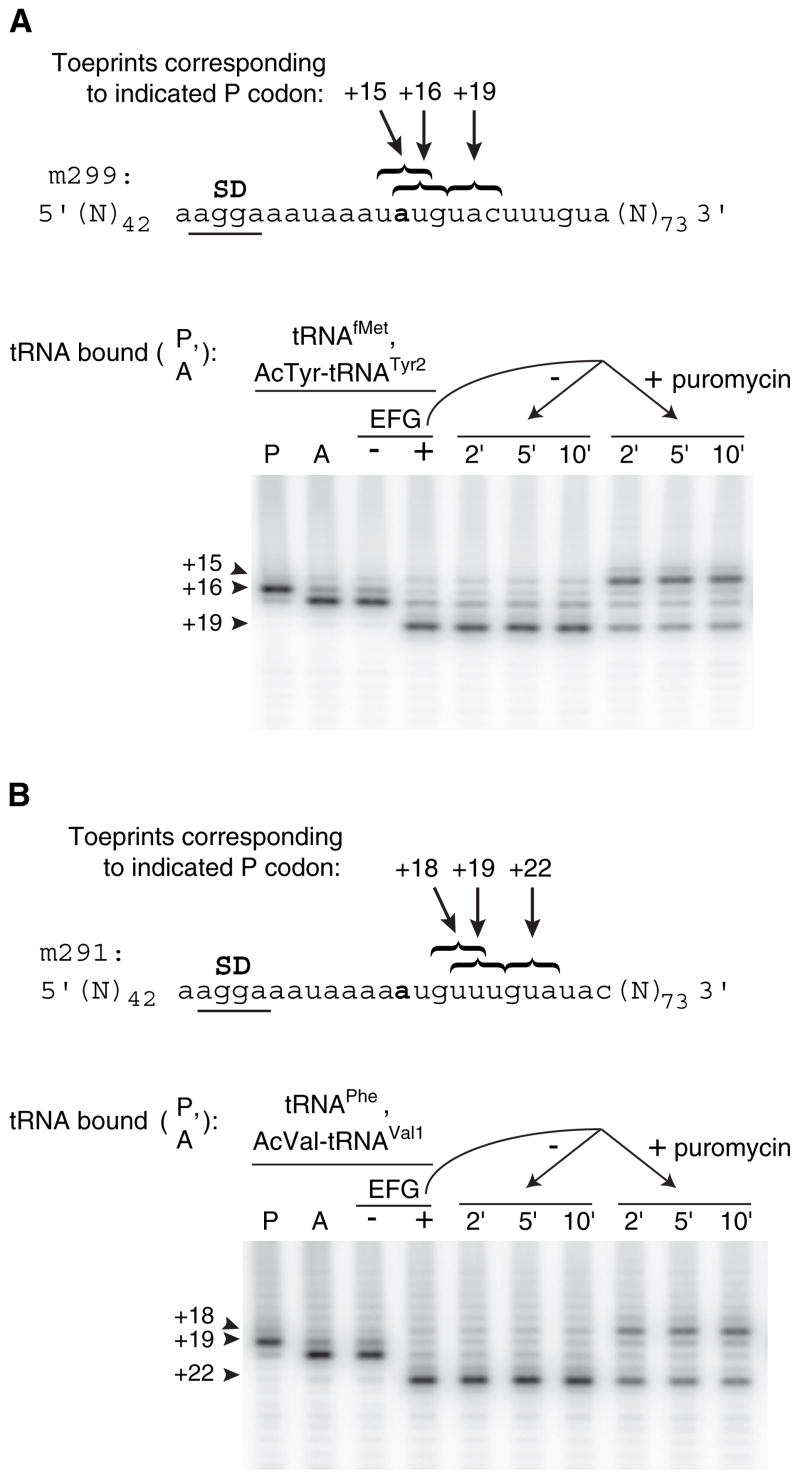
Figure 1. Deacylation of N-Acetyl-Aminoacyl-tRNA in the Posttranslocation Complex Results in mRNA Slippage The position of mRNA in ribosomal complexes was mapped by toe-printing after each of several additions.
(A) A pretranslocation complex was made by incubating ribosomes with message m299 and tRNAfMet to fill the P site (lane P) and then adding N-acetyl-Tyr-tRNATyr2 to fill the A site (lane A). Next, this pretranslocation complex was incubated with either GTP alone as a control (− lane) or with EF-G plus GTP to form the posttranslocation complex (+ lane). Finally, this posttranslocation complex was further incubated without or with addition of puromycin (1.7 mM) as indicated.
(B) In an analogous experiment, a posttranslocation complex was formed by translocation of N-acetyl-Val-tRNAVal1 paired to the GUA codon of m291, and slippage of the mRNA was observed after addition of puromycin.