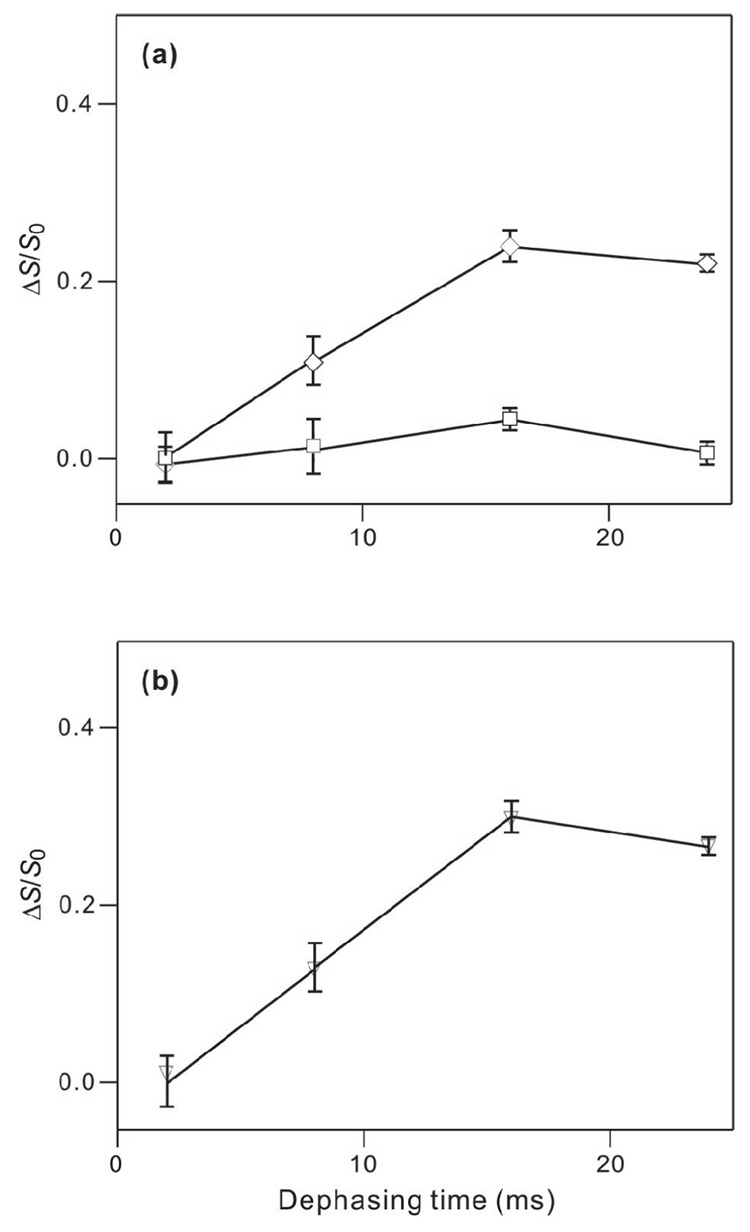
Figure 7.
(ΔS/S0) vs dephasing time for HFP4-6A/PC:PG:CHOL and HFP-14A/PC:PG:CHOL samples. In panel a, (ΔS/S0)exp points with error bars are displayed with legend: HFP4-6A, squares; and HFP4-14A, diamonds. Each (ΔS/S0)exp value was determined from integrals of the entire S0 and S1 peaks. Panel b represents analysis of the HFP4-14A data with legend: (ΔS/S0)lab, vertical lines with error bars; and best-fit (ΔS/S0)sim, down triangles. Lines are drawn between points with adjacent τ values. The best-fit parameters were d = 93 ± 10 Hz with corresponding r = 5.08 ± 0.19 Å, f = 0.29 ± 0.02, and χ2min = 0.1.