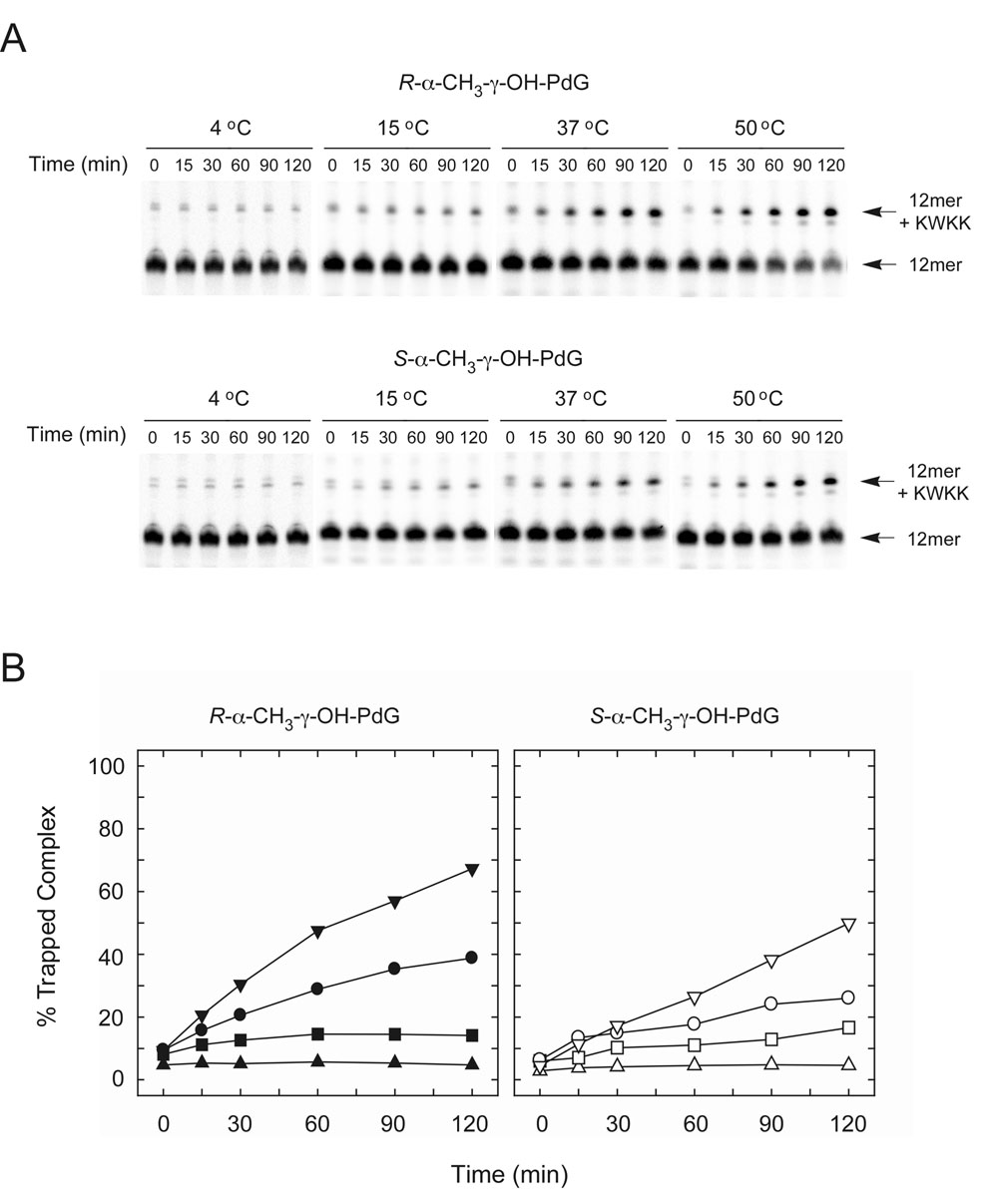
Figure 2.
DNA-peptide crosslinking involving R- and S-α-CH3-γ-OH-PdG adducts. A. For trapping reactions, single-stranded crotonaldehyde-adducted oligodeoxynucleotides (75 nM) were incubated with 1.0 mM KWKK the presence of 50 mM NaCNBH3 at 4, 15, 37 or 50 °C. Reactions were carried out in 100 mM HEPES (pH 7.0) and 100 mM NaCl and were incubated for 0, 15, 30, 60, 90 or 120 min. Reactions were quenched at the end of the incubation period by the addition of 100 mM NaBH4. Labels indicate the positions of the substrate 12-mer DNAs and the major reduced Schiff base conjugates (12-mer + peptide) following denaturing PAGE analysis. B. Kinetics of trapped conjugate formation are plotted over the 2 h time course at 4 °C [R-α-CH3-γ-OH-PdG, π; S-α-CH3-γ-OH-PdG, ρ], 15 °C [R-α-CH3-γ-OH-PdG, ´; S-α-CH3-γ-OH-PdG, ≤], 37 °C [R-α-CH3-γ-OH-PdG, ; S-α-CH3-γ-OH-PdG, ], and 50 °C [R-α-CH3-γ-OH-PdG, θ; S-α-CH3-γ-OH-PdG, σ].