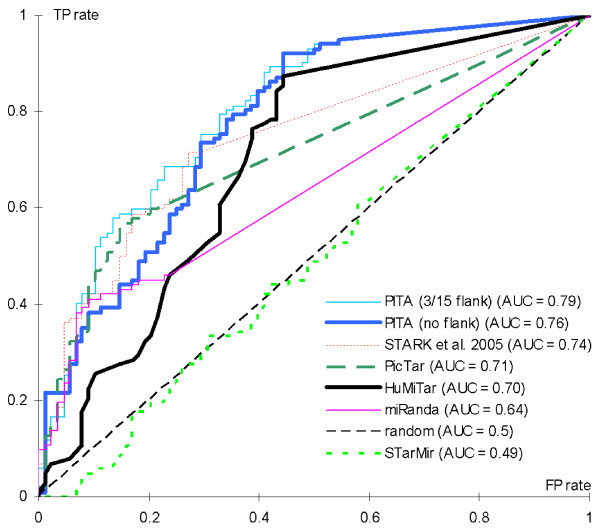
Figure 4.
ROC curves and the corresponding AUC scores that quantify sensitivity and specificity of different miR target predictors on the interactions set. The results include five existing prediction methods, PicTar [10], miRanda [19], method by Stark and colleagues [35], STarMir [36], and PITA [30]. The latter method includes two versions, one requiring unpairing of only target-site nucleotides (PITA no flank) and another that also requires unpairing of 3 and 15 flanking nucleotides upstream and downstream of the target site (PITA 3/15 flank), respectively. For each prediction method, the targets were sorted by score and the FP rates (x axis) and TP rates (y axis) were plotted for each possible score prediction threshold. The area under the curve (AUC) for each method is shown in the figure legend. The AUC is computed by extending each plot to the upper right corner as in [30]. The results obtained by a random sorting of the targets are shown using a thin dashed line. The ROC curves and AUC values of PITA, STARK, PicTar, miRanda, and STarMir were taken from [30].