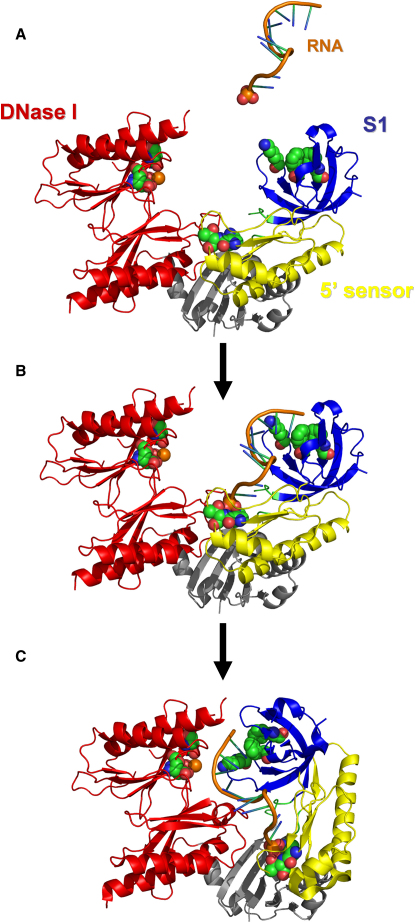
Figure 3.
Proposed Mechanism of Substrate Binding and Catalysis by RNase E
(A) In the absence of RNA, the monomer is in an open state in the highly dynamic apoprotein state. The S1 subdomain and 5′ sensing site are both exposed to the surrounding solvent, allowing RNA to readily bind.
(B) The 5′ sensing pocket likely contributes a significant portion of the substrate-binding affinity, with the S1 subdomain acting to orient the molecule.
(C) After RNA is bound, the consolidated 5/S1 subdomain moves as a unit in a conformation change that brings the substrate into close proximity to the catalytic site on the DNase I subdomain. The RNA is cleaved and the products are released as the structure returns to the open configuration. Depicted in space-filling representation are the key recognition residues of the 5′ sensing pocket and the S1 subdomain, the catalytic residues on the DNase I domain, and the 5′ phosphate of the single-stranded RNA substrate.