Abstract
The bacterial protein DsbD transfers reductant from the cytoplasm to the otherwise oxidizing environment of the periplasm. This reducing power is required for several essential pathways, including disulfide bond formation and cytochrome c maturation. DsbD includes a transmembrane domain (tmDsbD) flanked by two globular periplasmic domains (nDsbD/cDsbD); each contains a cysteine pair involved in electron transfer via a disulfide exchange cascade. The final step in the cascade involves reduction of the Cys103-Cys109 disulfide of nDsbD by Cys461 of cDsbD. Here we show that a complex between the globular periplasmic domains is trapped in vivo only when both are linked by tmDsbD. We have found previously (Mavridou, D. A., Stevens, J. M., Ferguson, S. J., & Redfield, C. (2007) J. Mol. Biol. 370 ,643 -658) that the attacking cysteine (Cys461) in isolated cDsbD has a high pKa value (10.5) that makes this thiol relatively unreactive toward the target disulfide in nDsbD. Here we show using NMR that active-site pKa values change significantly when cDsbD forms a complex with nDsbD. This modulation of pKa values is critical for the specificity and function of cDsbD. Uncomplexed cDsbD is a poor nucleophile, allowing it to avoid nonspecific reoxidation; however, in complex with nDsbD, the nucleophilicity of cDsbD increases permitting reductant transfer. The observation of significant changes in active-site pKa values upon complex formation has wider implications for understanding reactivity in thiol:disulfide oxidoreductases.
DsbD is a unique protein that transfers reductant across the cytoplasmic membrane to the periplasm in many Gram-negative bacteria (1, 2). Provision of reductant to the periplasm is required because this compartment is otherwise considered to be an oxidizing environment (2). DsbD includes three domains, each containing a pair of cysteine residues that perform a series of disulfide exchange reactions (Fig. 1A). In the first step, the transmembrane domain (tmDsbD) accepts electrons from thioredoxin in the cytoplasm; these are then transferred to the periplasmic C-terminal domain (cDsbD) and finally to the N-terminal domain (nDsbD), which is also located in the periplasm (3-5). nDsbD acts as a junction point for several pathways that require reductant, including the general disulfide isomerase system and the pathway that is thought to reduce the cysteine thiols of apocytochromes in the cytochrome c biogenesis pathway (6). In Gram-positive bacteria, CcdA, an integral membrane protein, and ResA, which has a thioredoxin fold, provide the reductant required for cytochrome c maturation (7).
FIGURE 1.
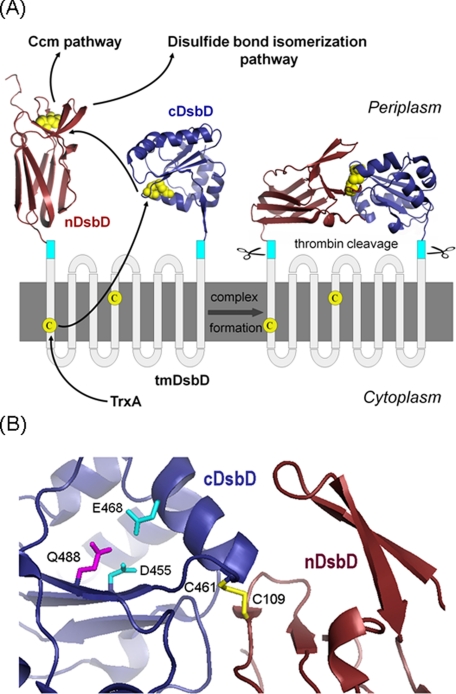
Schematic representation of DsbD. A, proposed pathway of electron flow from thioredoxin (TrxA) in the cytoplasm, via the three domains of DsbD, to the cytochrome c maturation (Ccm) and disulfide bond isomerization pathways in the periplasm is shown. The crystal structure of nDsbD is from Protein Data Bank code 1L6P (8), cDsbD from Protein Data Bank code 1UC7 (11), and the nDsbD-cDsbD complex from Protein Data Bank code 1VRS (12). The cyan boxes indicate the thrombin cleavage sites introduced into full-length DsbD to allow detection of the nDsbD-cDsbD complex following its formation in vivo. The cysteine residues are shown in yellow. B, schematic representation of the active site of cDsbD in the covalent complex with nDsbD (12). Some active-site residues of cDsbD are indicated in stick representation and the inter-domain disulfide (Cys461-SS-Cys109) is shown in yellow.
Structural studies have sought to explain how DsbD functions and interacts with its various partners. The structures of the two soluble periplasmic domains have been determined (Fig. 1A, left). nDsbD has an immunoglobulin-like structure (8, 9) and is the only known thiol:disulfide oxidoreductase with this fold. cDsbD has the more typical thioredoxin fold found in many oxidoreductases; this has the characteristic active-site CXXC motif (10, 11). A covalent complex between single-cysteine variants of each of these two domains was produced in vitro and its x-ray structure solved (12), revealing the interface between the two domains (Fig. 1A, right). Although this mixed disulfide is accepted as a physiological intermediate in the function of DsbD, an in vivo complex between the two soluble domains has not been reported previously (3). Further complexes between nDsbD and its other physiological partners have also been trapped and their structures examined (9, 13). Interestingly, all of the interaction partners of nDsbD are thioredoxin-like proteins; similarities in their folds are congruous with common interaction interfaces (14). However, only cDsbD will reduce nDsbD, whereas nDsbD will reduce several partners. This raises questions about how the direction of reductant flow is maintained and controlled within the series of disulfide-exchange reactions.
As part of our structural and mechanistic characterization of DsbD and its domains in solution, we have previously measured by NMR the pKa values of the active-site cysteine pair, Cys461 and Cys464, of cDsbD (numbered according to the full-length Escherichia coli DsbD sequence) (15). An unusually high pKa value of 10.5 was measured for the N-terminal cysteine of the CXXC motif, Cys461, and the pKa value of the second cysteine, Cys464, was significantly higher than the maximum pH value that was studied (pH 12.2). The pKa value of 10.5 is the highest reported for the N-terminal cysteine of the CXXC motif in a thioredoxin fold. The striking consequence of the elevated pKa value is that the active-site cysteine of cDsbD, Cys461, is not strongly nucleophilic, raising critical questions about how this cysteine reacts with the disulfide in nDsbD. It was demonstrated using site-directed mutagenesis that the negatively charged side chains of Asp455 and Glu468, which are located close to the CXXC motif (Fig. 1B), are responsible for the unusually high pKa value of Cys461; mutation of one or both of these residues to Asn and Gln, respectively, resulted in decreases in the pKa value of Cys461 from 10.5 to 9.9 (E468Q), to 9.3 (D455N), and to 8.6 (D455N/E468Q). The pKa values for Asp455 were found to be 5.9 and 6.6 in oxidized and reduced cDsbD; these values are significantly higher than the value of ∼4 for an unperturbed aspartic acid. We postulated that the properties of the amino acid side chains in the immediate environment of the cysteines in cDsbD would change upon complex formation with nDsbD, changing the reactivity of the cysteines and explaining how the reaction between the two domains is initiated (15). Specifically, we proposed that an increase in the pKa value of Asp455 upon complex formation would lead to a decrease in the pKa value of Cys461, thereby making it a better nucleophile. Stirnimann et al. (10) previously presented pKa calculations suggesting an increase in the Asp455 pKa value upon complex formation.
The aim of this work has been to determine the molecular basis of the control of the reactivity of the active-site cysteine residues in cDsbD, using NMR to compare the active-site properties of cDsbD alone and in its physiological complex with nDsbD. We demonstrate that the pKa value of Asp455 is elevated by at least 1.1 pH units when cDsbD forms a complex with nDsbD. This modulation of the pKa value is critical for the specificity and function of cDsbD. These in vitro studies are complemented by in vivo studies on complex formation, in which we have trapped the nDsbD-cDsbD complex for the first time. The results of our experiments explain how the intramolecular disulfide cascade within the soluble domains of DsbD functions, and demonstrate the importance of the transmembrane domain in controlling and facilitating complex formation between the soluble domains.
EXPERIMENTAL PROCEDURES
Construction of DsbD Plasmids—The plasmid pDzc1 was used to express isolated wild-type cDsbD bearing a C-terminal His6 tag and the plasmid pDzc5 a single cysteine variant (C464A) of cDsbD with a thrombin cleavage site preceding the C-terminal His6 tag. Isolated nDsbD with the PelB signal peptide replacing the endogenous signal peptide and with a C-terminal streptavidin tag was expressed from the plasmid pDzn1, and from that a single-cysteine variant (C103A) was produced. Finally, the full-length construct in which nDsbD and cDsbD could be cleaved from tmDsbD (thrombin-cleavable DsbD) was expressed from pDsbd4. Details of the production of the above plasmids can be found in supplemental material.
Protein Production, Purification, and Characterization—All experiments were performed in the E. coli strain BL21(DE3) (Stratagene). All cells expressing unlabeled protein were grown at 37 °C in 500-ml volumes of Luria Bertani (LB) broth in 2-liter flasks from overnight starter cultures (grown at 37 °C). SDS-PAGE analysis was carried out on 10-20% BisTris6 NuPAGE gels (Invitrogen) with prestained protein markers (SeeBlue Plus 2, Invitrogen).
Overexpression of Thrombin-cleavable DsbD—Bacteria transformed with pDsbd4 were grown with 100 μg ml-1 carbenicillin to an A600 of 0.7 before addition of 0.5 mm isopropyl β-d-thiogalactopyranoside. After further incubation for 3 h the cells were harvested, and the crude membrane fraction was isolated using a French press. Disruption of the cells was performed at 16,000 p.s.i. which was followed by centrifugation at 257,000 × g for 1.5 h at 4 °C. The membrane fraction was solubilized at a protein concentration of 5 mg ml-1 for 1 h at 4 °C under gentle agitation in 20 mm Tris-HCl, 300 mm NaCl, 20% v/v glycerol, 1% w/v n-dodecyl β-d-maltoside (DDM) (Anatrace) (pH 7.5). Unsolubilized material was removed by centrifugation at 257,000 × g for 45 min at 4 °C. The supernatant was applied to 10 ml of Fast Flow Chelating-Sepharose (Amersham Biosciences) charged with Ni2+. The column was washed with 20 mm Tris-HCl, 150 mm NaCl, 20 mm imidazole, 0.1% w/v DDM (pH 7.5), and the bound protein was eluted with 20 mm Tris-HCl, 150 mm NaCl, 200 mm imidazole, 0.1% w/v DDM (pH 7.5). A concentrated protein solution of 0.5 ml was applied to a Superdex 200 (HR) size-exclusion column (GE Healthcare) equilibrated in 20 mm Tris-HCl, 150 mm NaCl, 0.03% w/v DDM (pH 7.5). The purest fractions were pooled and concentrated to 0.5 ml. To prevent artifactual thiol-disulfide exchange reactions from occurring during the thrombin cleavage, free thiols were alkylated with N-ethylmaleimide (Sigma) (16). Purified protein was incubated in the presence of 30 mm N-ethylmaleimide for 1 h at room temperature. After removal of the excess alkylating reagent, the protein was cleaved using the thrombin Clean-Cleave kit according to the manufacturer's instructions (Sigma). The presence of the nDsbD-cDsbD mixed disulfide was confirmed by Western blotting using two primary antibodies as follows: 1) penta-His HRP-conjugated monoclonal antibody (Qiagen), and 2) goat antiserum raised against the cDsbD sequence of E. coli DsbD and donkey anti-sheep alkaline phosphatase-conjugated antibody (Sigma), as primary and secondary antibodies, respectively.
Characterization of the nDsbD-cDsbD Mixed Disulfide Formed in Vivo—The covalent complex between nDsbD and cDsbD was N-terminally sequenced from protein samples in SDS-polyacrylamide gels that were electrophoretically transferred to a polyvinylidene difluoride membrane and stained with Coomassie Brilliant Blue. The protein bands were excised and subjected to automated Edman sequencing using an Applied Biosystems 494A Procise protein sequencer.
Simultaneous Overexpression of nDsbD and C464A-cDsbD—Bacteria transformed with pDzn1 and pDzc5 were grown with 100 μg ml-1 ampicillin and 20 μg ml-1 gentamycin to an A600 of 1.2 before addition of 1 mm isopropyl β-d-thiogalactopyranoside. After further incubation for 4 h, the cells were harvested spheroplasted as described (17) omitting EDTA. Western blotting of the extracted periplasm for the detection of the nDsbD-cDsbD mixed disulfide complex was done using two primary antibodies as follows: 1) StrepMAB-Classic HRP-conjugated monoclonal antibody (IBA GmbH), and 2) penta-His HRP-conjugated monoclonal antibody (Qiagen).
Production and Purification of the C103A-nDsbD-15N-C464A-cDsbD Complex for NMR Studies—The mixed disulfide complex was formed in vitro using C103A-nDsbD and uniformly 15N-labeled C464A-cDsbD purified separately from BL21(DE3) cells (Stratagene). For the production of C103A-nDsbD, cells transformed with pDzn2 were grown at 30 °C with 20 μg ml-1 gentamycin to an A600 of 1.5 before addition of 1 mm isopropyl β-d-thiogalactopyranoside. After further incubation for 4 h at 37 °C the cells were harvested and spheroplasted as described (17) omitting EDTA. The periplasmic fraction was applied to 5 ml of Strep-Tactin-Sepharose (IBA GmbH) equilibrated with 50 mm Tris-HCl, 150 mm NaCl (pH 7.5). The column was washed with 50 mm Tris-HCl, 1 m NaCl (pH 7.5), and the protein was eluted with 50 mm Tris-HCl, 150 mm NaCl, 2.5 mm desthiobiotin (IBA GmbH) (pH 7.5) according to the manufacturer's instructions. Production and purification of 15N-labeled C464A-cDsbD were done in the same way as for wild-type cDsbD, as described in previous work (15).
The C103A-nDsbD-15N-C464A-cDsbD mixed disulfide complex was prepared using a reported method (12). First, C103A-nDsbD was mixed with 10 mm dithiobisnitrobenzoic acid (Sigma) at 25 °C for 30 min. After removal of excess dithiobisnitrobenzoic acid, it was mixed with equimolar 15N-C464A-cDsbD and incubated as above. The reaction mixture was purified by two affinity chromatography steps, each specific to one of the domains. The mixture was applied to 3 ml of Fast Flow Chelating-Sepharose (Amersham Biosciences) charged with Ni2+and purified as described previously (15). After removing the imidazole, the protein solution was applied to 3 ml of Strep-Tactin-Sepharose (IBA GmbH) and was purified as described above for C103A-nDsbD. The nDsbD-15N-cDsbD mixed-disulfide sample was of high purity as assessed using SDS-PAGE (supplemental Fig. S1A), and its correct molecular weight was verified by electrospray ionization-mass spectrometry (expected mass, 33,187 Da; observed mass, 33,185 Da).
Production and Purification of the 15N-labeled Wild Type and C464A-cDsbD—15N-Labeled wild-type cDsbD and C464A-cDsbD were used as control proteins for the pH titration of the nDsbD-15N-cDsbD mixed disulfide. Both proteins were produced and purified as described previously (15).
pH Titration of the nDsbD-15N-cDsbD Mixed Disulfide Using NMR Spectroscopy—The 1HN and 15N resonances of C464A-cDsbD as a separate domain and in the nDsbD-15N-cDsbD mixed disulfide were assigned using uniformly 15N-labeled samples of 0.8 mm protein for the isolated domains and 0.5 mm protein for the mixed disulfide, in 95% H2O, 5% D2O at pH 6.5. Assignments for C464A-cDsbD in the isolated domain and in the nDsbD-15N-cDsbD complex were obtained by comparison of three-dimensional 15N-edited nuclear Overhauser effect spectroscopy-HSQC and total correlation spectroscopy-HSQC spectra with the previously assigned spectra of reduced wild-type cDsbD (15).
NMR experiments for the determination of the pKa values of Asp455 in the active-site of cDsbD in the nDsbD-15N-cDsbD mixed disulfide were performed using 0.5 mm of complex in 95% H2O, 5% D2O. Determination of the pKa values of Asp455 in the active-site of 15N-labeled wild-type cDsbD and C464A-cDsbD was done in the same way as a control experiment. In the case of the 15N-labeled wild-type cDsbD, the sample was a ∼1:1 mixture of oxidized and reduced protein allowing simultaneous measurement of the pKa value for the two oxidation states. The pH of solutions was adjusted by using small volumes of 0.1-1 m HCl or 0.1-1 m NaOH. The pH of the samples was measured before and after each experiment, and the average of the two measurements was used for data analysis. The pH values given are direct pH meter readings measured at room temperature. Two-dimensional 1H-15N HSQC spectra were collected at 313 K on a home-built 750-MHz NMR spectrometer, which is controlled with GE/Omega software and is equipped with a home-built triple-resonance pulsed-field gradient probe-head. Sweep widths of 9345.79 Hz and 2500 were used in F2 (1H) and F1 (15N), respectively. 128 complex 15N increments of 8, 12, and 96 scans were collected with 1024 complex points in the acquisition dimension for 15N-labeled wild-type cDsbD, C464A-cDsbD, and the nDsbD-15N-cDsbD mixed disulfide complex, respectively. NMR data were processed and visualized using NMRPipe, NMRDraw, and NMRView (18, 19). Peak picking was carried out using in-house software.
pH Titration of [3-13C]Cysteine-labeled Wild Type and D455N-cDsbD—1H-13C HSQC experiments for the determination of the pKa value of Cys461 at 313 K were performed on solutions containing ∼1.5 mm of [3-13C]cysteine-labeled wild-type or D455N cDsbD in 99% D2O as described previously at 298 K (15). Dithiothreitol was added to maintain the proteins in the reduced state.
Data Fitting—pKa values for Asp455 were determined from 1HN or 15N chemical shifts measured as a function of pH. pKa values for Cys461 were determined from 1Hβ or 13Cβ chemical shifts measured as a function of pH. The titration data were fitted to either one or two pKa curves (20) using in-house software as described previously (15). The pKa value for Asp455 was determined from the pH dependence of five chemical shift values (Asp455/Glu468 1HN and 15N and Gln488 15Nε). The pKa value for Cys461 was determined from the pH dependence of four to six chemical shift values (Cys461/Cys464 1Hβ and 13Cβ). Errors in the pKa values were estimated as the standard deviation from the mean pKa value. Details of the parameters obtained in each fit are listed in supplemental Tables S1 and S2.
RESULTS
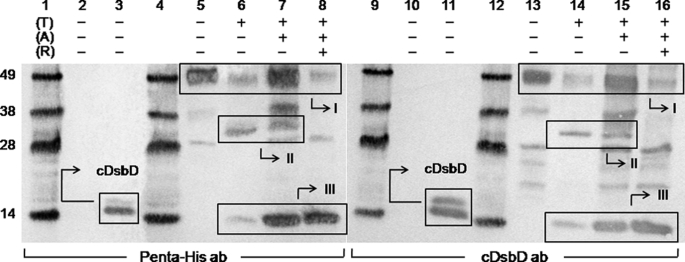
In Vivo Studies of the Formation of an nDsbD-cDsbD Complex in Full-length DsbD—We have used a thrombin-cleavable variant of DsbD (Fig. 1A) to attempt to identify a mixed-disulfide complex of nDsbD and cDsbD in vivo. This construct expresses full-length DsbD in which nDsbD and cDsbD can each be cleaved from the transmembrane domain by virtue of engineered thrombin cleavage sites. The C464A mutation in cDsbD was introduced in an attempt to trap the nDsbD-cDsbD mixed-disulfide complex because Cys464 is believed to be responsible for driving the cleavage of the interdomain disulfide and the final transfer of reductant from cDsbD to nDsbD (12). Thrombin-cleavable DsbD was expressed, purified, incubated with N-ethylmaleimide to alkylate its free thiols, incubated with thrombin, and analyzed by SDS-PAGE and Western blotting with antibodies against the His6 tag and cDsbD; the results of the Western blotting are shown in Fig. 2. It can be seen in Fig. 2, lanes 6, 7, 14, and 15, that a band of the expected molecular weight for an nDsbD-cDsbD mixed disulfide (∼32 kDa) appeared after thrombin cleavage of thrombin-cleavable DsbD (box II). No band can be seen at this position before thrombin cleavage (Fig. 2, lanes 5 and 13). The ∼32-kDa band disappears in the presence of reductant (Fig. 2, lanes 8 and 16), confirming that it is a covalent complex containing a disulfide bond. The ∼32-kDa covalent complex from the SDS-PAGE band was N-terminally sequenced, and both the nDsbD ((M)LFDAPGRSQ) and cDsbD (GSGQTHLNFT) termini were detected. As seen in Fig. 2, lanes 6 and 7 or 14 and 15, the band corresponding to the complex is present whether or not the free thiols of the protein were alkylated before thrombin cleavage. Incubation with N-ethylmaleimide, therefore demonstrates that this complex was formed before cleavage of the full-length protein. A band for the full-length thrombin-cleavable DsbD can be seen (at ∼49 kDa) even after thrombin cleavage (Fig. 2, lanes 6-8 and 14-16, box I); this is attributed to inefficient thrombin cleavage because of the presence of the detergent DDM in the protein solutions. It is also likely that the presence of the protein in the detergent micelle could reduce the accessibility of the protease cleavage sites to the thrombin (21). Moreover, a C464A-cDsbD band can be seen (at ∼14 kDa) after thrombin cleavage even in the absence of reductant (Fig. 2, lanes 6, 7, 14, and 15, box III). This is observed because there are molecules in the protein sample in which the nDsbD-cDsbD mixed disulfide had not formed and for which the thrombin cleavage led to release of the two separate periplasmic domains (isolated nDsbD cannot be seen in the Western blot because it is not His6-tagged).
FIGURE 2.
Western blots of purified full-length C464A thrombin-cleavable DsbD before and after thrombin cleavage. Penta-His HRP-conjugated antibody was used in lanes 1-8 and goat antiserum raised against cDsbD of E. coli DsbD was used in lanes 9-16. Lanes 1, 4, 9, and 12 show molecular mass markers (from the top these are 49, 38, 28, and 14 kDa); lanes 2 and 10 show the negative control (nDsbD with no affinity tag); lanes 3 and 11 show the positive control (cDsbD bearing a C-terminal His6 tag). C464A thrombin-cleavable DsbD is shown in lanes 5 and 13 before alkylation of free thiols (A) and thrombin cleavage (T), in lanes 6 and 14 without alkylation and after thrombin cleavage, in lanes 7 and 15 after alkylation and thrombin cleavage, and in lanes 8 and 16 after alkylation and thrombin cleavage in the presence of reductant (R). The cDsbD band can be seen at ∼14 kDa in lanes 3 and 11, and in 6-8 and 14-16 in Box III (this construct runs as a diffuse double band on SDS-PAGE because its PelB signal sequence, which targets the protein to the periplasm, is cleaved inefficiently by the signal peptidase, leaving a large fraction of the protein uncleaved (confirmed by mass spectrometry and N-terminal sequencing)). The uncleaved C464A thrombin-cleavable DsbD band can be seen at ∼49 kDa (the actual mass of the protein is 60.8 kDa; a difference between the two masses is common for membrane proteins) in lanes 5-8 and 13-16 (box I) along with another band at ∼28 kDa that is a contaminant commonly seen after the purification of this protein. The nDsbD-cDsbD mixed disulfide band can be seen at ∼32 kDa in lanes 6, 7, 14, and 15 (box II) and it disappears in the presence of reductant in lanes 8 and 16. 2-3 μg of total protein were loaded in each lane.
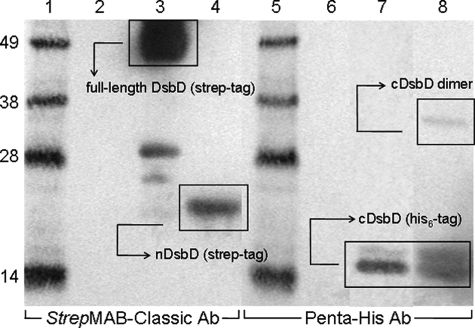
In Vivo Studies of Complex Formation between Isolated nDsbD and cDsbD—To assess the role of tmDsbD in mediating the interaction between nDsbD and cDsbD, the two periplasmic domains of DsbD, nDsbD bearing a C-terminal streptavidin tag and a C464A variant of cDsbD bearing a C-terminal His6 tag, were overexpressed simultaneously as soluble proteins exported to the periplasm. The C464A mutation in cDsbD was used for reasons described above. The periplasmic extract was analyzed by SDS-PAGE and Western blotting with antibodies against the streptavidin tag, to detect nDsbD, and against a penta-histidine sequence, to detect cDsbD. The nDsbD-cDsbD mixed disulfide is expected to migrate on SDS-PAGE between the 28- and 38-kDa molecular mass markers and to be detected by both antibodies (Fig. 3). No band corresponding to the nDsbD-cDsbD mixed disulfide was observed in Fig. 3, lanes 4 and 8. In Fig. 3, lane 4, only the nDsbD band at ∼17 kDa is observed, and in lane 8 the C464A-cDsbD band at ∼14 kDa and a band corresponding to its homodimer at ∼33 kDa (confirmed by Western blotting, not shown) are observed.
FIGURE 3.
Western blots of periplasmic extracts of E. coli expressing nDsbD with a streptavidin tag and C464A-cDsbD with a His6 tag. StrepMAB-Classic HRP-conjugated antibody was used in lanes 1-4 and penta-His HRP-conjugated antibody was used in lanes 5-8. Lanes 1 and 5 show molecular mass markers; lanes 2 and 6 show the negative control (nDsbD with no affinity tag); lanes 3 and 7 show positive controls (full-length DsbD with a C-terminal streptavidin tag and cDsbD with a C-terminal His6 tag, respectively), and lanes 4 and 8 show the periplasmic extract. The full-length DsbD band can be seen at ∼49 kDa in lane 3 along with a small amount of contaminant at ∼28 kDa (see explanation in Fig. 2). The nDsbD band can be seen at ∼17 kDa in lane 4 and the cDsbD band can be seen at ∼14 kDa (the diffuse double band is explained in Fig. 2) in lanes 7 and 8 along with the band of its homodimer at ∼33 kDa. 15-20 μg of total protein were loaded in each lane.
A possible explanation for the failure of the nDsbD-cDsbD mixed-disulfide complex to form in vivo is that the proteins were present in the incorrect oxidation states for disulfide bond formation to occur; interdomain disulfide bond formation requires the presence of oxidized nDsbD and reduced C464A-cDsbD. NMR studies of periplasmically overexpressed nDsbD show that it is present predominantly in the oxidized form. cDsbD cannot form an intramolecular disulfide bond because it has a single cysteine and the homodimer is observed as only a minor species (Fig. 3, lane 8). For these reasons, attack of the cDsbD cysteine thiol on the disulfide of nDsbD, as occurs in the physiological pathway, should have been possible. We therefore conclude that a complex between soluble nDsbD and cDsbD, co-expressed in the E. coli periplasm, does not form in the absence of the transmembrane domain that usually connects the two domains in the full-length protein.
NMR Analysis of the nDsbD-15N-cDsbD Mixed-disulfide Complex—A mixed-disulfide complex was produced using purified single-cysteine variants of nDsbD (C103A) and cDsbD (C464A) to ensure that the physiologically relevant disulfide was formed (Cys461-SS-Cys109), to avoid any cleavage of this disulfide because of the presence of Cys464 and to eliminate any complications during the pKa determination of Asp455 caused by the titration of the Cys464 or Cys103 thiols in the active site. NMR spectroscopy allows measurement of the pKa value of Asp455 in the active site of cDsbD when cDsbD is in close proximity to its periplasmic partner nDsbD. For this purpose we used uniformly 15N-labeled C464A-cDsbD covalently linked to unlabeled C103A-nDsbD, thus producing a C103A-nDsbD-15N-C464A-cDsbD covalent complex (subsequently referred to as nDsbD-15N-cDsbD) where only cDsbD gives NMR signals in the 1H-15N HSQC spectrum but in which it is influenced by the presence of nDsbD. In this way we were able to simplify the NMR spectrum of a relatively large (∼32-kDa) protein complex and to focus on the domain of interest.
Previous studies of isolated cDsbD were carried out at 298 K. The 1H-15N HSQC spectrum of the nDsbD-15N-cDsbD complex at 298 K is significantly broader, because of the higher molecular weight of the complex, than that of isolated cDsbD, and many of the expected peaks are not observed (supplemental Fig. S1, D and E). A significant improvement in the quality of the HSQC spectrum of the complex was observed at 313 K (supplemental Fig. S1F); at this temperature all expected peaks from cDsbD are observed. Both nDsbD and cDsbD have relatively high thermal stabilities and are completely folded and native at 313 K (supplemental Fig. S1B). The pH titration experiments described below were therefore performed at 313 K.
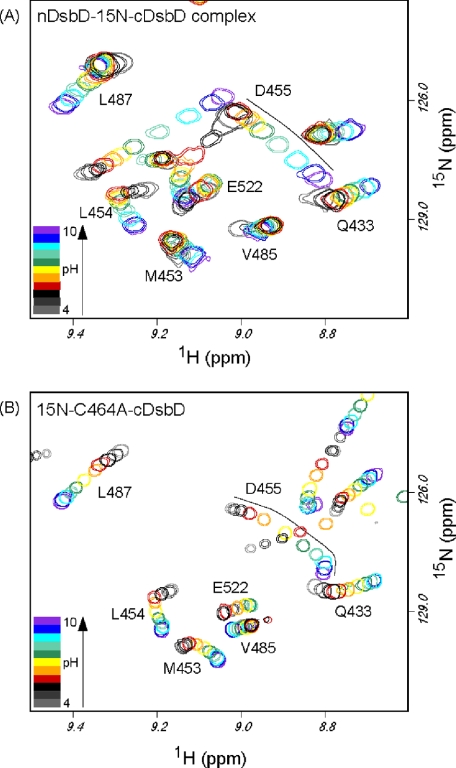
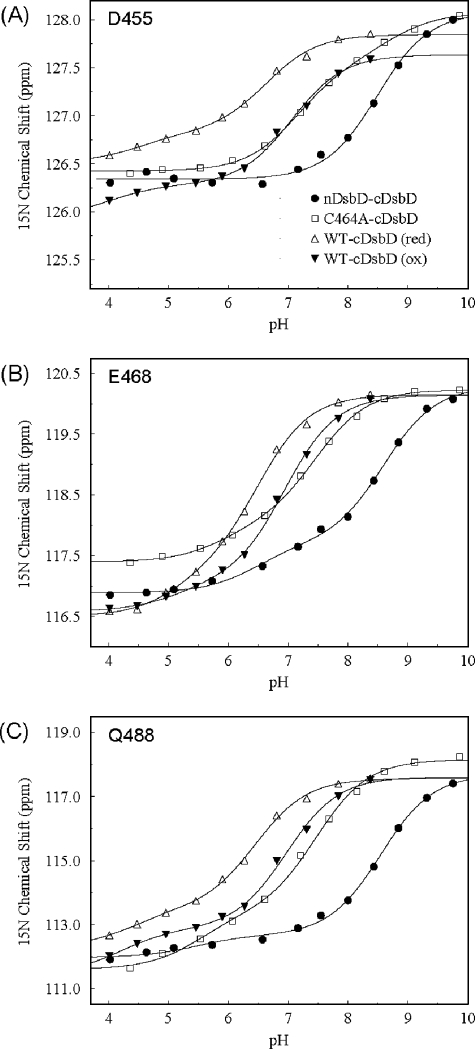
pKa Determination for Asp455 in the nDsbD-15 N-cDsbD Complex—We have monitored, between pH 4 and pH 10, the 15N and 1HN chemical shifts of the backbone amides of Asp455 and Glu468 and the side chain 15Nε of Gln488 in HSQC spectra of the nDsbD-15N-cDsbD mixed-disulfide complex (Fig. 4A). These shifts are all sensitive to titration of Asp455 because of a hydrogen bond network involving Asp455, Glu468, and Gln488 (10, 11). We have shown previously that cDsbD is stable between pH 4 and 12 (15). 1H NMR spectra of the complex collected between pH 4 and 10 demonstrate that nDsbD in the complex is also stable over this pH range (supplemental Fig. S1C). Analysis of the pH dependence of the 15N and 1HN chemical shifts of Asp455 and Glu468 and of the side chain 15Nε of Gln488 gave an average pKa value of 8.5 ± 0.1 for Asp455 (Fig. 5, A-C). This pKa value is significantly higher than the values of 5.9 ± 0.1 and 6.6 ± 0.1 obtained previously for Asp455 in reduced and oxidized cDsbD (15). Our previous study of cDsbD was carried out at 298 K, whereas the spectra of the complex were collected at 313 K. As a control, we have monitored the same chemical shifts in uniformly 15N-labeled wild-type cDsbD at 313 K. The experiment was performed, as before (15), in a mixed oxidation state sample, and we obtained a pKa value of 6.5 ± 0.1 in the reduced state and of 7.0 ± 0.1 for Asp455 in the oxidized state (Fig. 5, A-C). These values are ∼0.5 pH units higher than the values measured at 298 K but are nevertheless still significantly lower than the value of 8.5 measured for Asp455 in the nDsbD-15N-cDsbD complex. To ensure that the difference in the pKa values of Asp455 between complexed and uncomplexed cDsbD is not an effect of the C464A mutation, we have also performed the titration for uniformly 15N-labeled C464A-cDsbD at 313 K and obtained an average pKa value of 7.4 ± 0.1 for Asp455 (Fig. 4B and Fig. 5, A-C). Therefore, it can be concluded that the pKa value of Asp455 is elevated by at least 1.1 pH units when cDsbD forms a complex with nDsbD.
FIGURE 4.
Overlay of two-dimensional 1H-15N HSQC spectra of the nDsbD-15N-C464A-cDsbD complex (A) and 15N-C464A-cDsbD collected at 750 MHz and 313 K for pH values ranging from 4 to 10 (B). For the nDsbD-15N-C464A-cDsbD complex, spectra collected at pH 4.635 (light gray), 5.09 (dark gray), 5.73 (black), 6.57 (red), 7.165 (orange), 7.55 (yellow), 8.00 (green), 8.43 (blue-green), 8.845 (cyan), 9.32 (blue), and 9.755 (violet) are shown. For 15N-C464A-cDsbD, spectra collected at pH 4.36 (light gray), 4.895 (dark gray), 5.53 (black), 6.075 (red), 6.61 (orange), 7.21 (yellow), 7.685 (green), 8.155 (blue-green), 8.61 (cyan), 9.11 (blue), and 9.86 (violet) are shown. The black lines drawn in A and B trace the pH-dependent chemical shift changes observed for Asp455.
FIGURE 5.
Measurement of the pKa value of the active-site residue Asp455 from the pH dependence of the 15N chemical shift of Asp455 (A), the 15N chemical shift of Glu468 (B), and the 15Nε chemical shift of Gln488 (C). Plots are shown for the nDsbD-C464A-cDsbD complex (•), isolated C464A-cDsbD (□), and for oxidized (▾) and reduced (▵) isolated wild-type cDsbD. The continuous lines show the best fit to a single pKa value or to two pKa values for each dataset. The fitting procedure has been described previously (15); see supplemental Table S1 for a summary of the fitted parameters.
pKa Determination for Cys461 at 313 K—The pKa value for Cys461 was measured for wild-type cDsbD and for the D455N variant of cDsbD at 313 K for direct comparison with the pKa values measured above for Asp455 in the nDsbD-15N-cDsbD complex at 313 K. Analysis of the pH dependence of the 13Cβ and 1Hβ chemical shifts of Cys461 and Cys464 in cDsbD gives pKa values of 10.4 ± 0.1 and 9.2 ± 0.2 for Cys461 in wild-type and D455N cDsbD, respectively (supplemental Fig. S2 and supplemental Table S2). These values agree within experimental error with the values of 10.5 ± 0.1 and 9.3 ± 0.05 reported previously at 298 K. Therefore, the pKa value of Cys461 does not show a temperature dependence.
DISCUSSION
It has long been known that DsbD functions by interaction between the cysteine pairs in its three domains, although how these interactions occur within this protein, in which one of the three domains is membrane-embedded, has remained elusive. In this study we have used both in vivo and in vitro approaches to understand how the interaction and reaction between the two soluble domains are controlled. We have demonstrated using Western blotting that a covalent nDsbD-cDsbD complex can be trapped in vivo when full-length DsbD is overexpressed. This result proves that the nDsbD-cDsbD interaction, which has previously only been probed in vitro (12), is physiologically relevant. No such complex was detected when the individual domains, nDsbD and cDsbD, were co-expressed in the periplasm.
The fact that the nDsbD-cDsbD complex was not detectable in the absence of the linking tmDsbD has important implications for the function of the membrane-embedded domain. The latter is known to interact with, and acquire reductant from, cytoplasmic thioredoxin and so effectively performs the transmembrane reductant transfer reaction (22). In addition, it appears to have an important function in ensuring the proximity and correct orientation of the two periplasmic domains. It should be noted that the interaction interface identified in the nDsbD-cDsbD crystal structure is not very extensive and involves a relatively small number of interactions (12). This limited interface may result in a relatively low affinity interaction between nDsbD and cDsbD. Therefore, the transmembrane domain may be essential to ensure the high local concentration of the domains that would be required for the formation of a complex and subsequent electron transfer via the disulfide exchange reaction.
In isolated cDsbD, Cys461, which catalyzes reduction of the Cys103-Cys109 disulfide in nDsbD, has a pKa value of 10.4 at 313 K; this elevated value makes Cys461 a poor nucleophile. The pKa value of Cys461 must therefore be lowered when it approaches nDsbD to make it more reactive. The high pKa value for Cys461 results from its close proximity to Asp455 and Glu468 in the active site. Here we have used the covalent nDsbD-15N-cDsbD complex to measure the pKa value of Asp455 in cDsbD when it is in close contact with nDsbD. We found that the pKa value of Asp455 is elevated by at least 1.1 pH units to a value of 8.5 in the complex at 313 K. At pH 7.0, the ratio of deprotonated (negatively charged) to protonated (uncharged) Asp455 in the complex will be 0.03:1. In effect, the carboxyl side chain of Asp455 is fully protonated. This will have a similar effect as the D455N mutation that we have found previously leads to a 1.2 pH unit decrease in the pKa value of Cys461 (from 10.4 to 9.2 at 313 K) (15). Therefore, we conclude that when nDsbD and cDsbD form a complex, a reduction in the pKa of Cys461 by at least 1.2 pH units to a value of ∼9.2 will take place. The concentration of the thiolate form of Cys461 will increase ∼16-fold at physiological pH making this cysteine a better nucleophile.
This experimental result is broadly consistent with previous electrostatic calculations carried out using x-ray structures, which predicted pKa values of 4.8 and 9.9 for Asp455 in the isolated cDsbD and the nDsbD-cDsbD complex, respectively (10). It should be noted that such calculations are extremely sensitive to small differences in x-ray structures; the program PROPKA gives predicted pKa values for Asp455 of 9.6, 7.0, and 7.7 using the three nDsbD-cDsbD molecules in the asymmetric unit (23). A pKa value of 5.4 is predicted by PROPKA for Asp455 in isolated reduced cDsbD. The increase in the predicted pKa value for Asp455 in the complex arises from an increase in the desolvation of the side chain of Asp455 and from an increased charge-charge contribution from Glu468 as this residue becomes more buried in the complex (23).
The side chains of Cys461 and Asp455 are in close proximity in the active site of cDsbD. These residues are homologous to Cys32 and Asp26 in thioredoxin; microscopic pKa values of 7.5 and 9.2 must be invoked to explain their titration behavior in thioredoxin (24, 25). If Asp26 titrates first with a pKa of 7.5, then Cys32 in the same molecule will have a pKa value of 9.2. Conversely, if Cys32 titrates with a pKa of 7.5, then Asp26 will have an elevated pKa of 9.2. In isolated and reduced cDsbD microscopic pKa values are not relevant because the measured pKa values of 6.5 and 10.4 for Asp455 and Cys461, respectively, at 313 K differ so significantly (15). However, when cDsbD comes into contact with nDsbD, and the pKa value of Asp455 is raised to 8.5, microscopic pKa values may need to be considered. In this case, if Asp455 titrates first with a pKa of 8.5, then Cys461 in the same molecule will have a pKa of ∼10.4. However, if Cys461 titrates first it will have a pKa of 8.5, and Asp455 will have a pKa of ∼10.4. In the latter case, Cys461 is significantly more reactive than in isolated cDsbD (∼80-fold increase in the concentration of the thiolate form of Cys461). This proposed lowering of the Cys461 pKa cannot be confirmed experimentally by NMR because nDsbD and cDsbD do not form at stable high affinity complex in the absence of a mixed disulfide involving Cys461.
The elevated pKa value of 10.4 for Cys461 in isolated cDsbD has the advantage of protecting cDsbD from nonspecific reoxidation, for example, by the powerful periplasmic oxidase DsbA. When cDsbD and nDsbD interact, the pKa value of Cys461 is lowered leading to an increase in the nucleophilic character of this residue. Asp455 is only able to exert this control over the reactivity of Cys461 in cDsbD because it is buried and already has a relatively high pKa value (∼6) in the isolated domain. Thus, modulation of active-site pKa values resulting from changes in the environment of the active site is key to the specificity of cDsbD.
Similar perturbations to pKa values may occur in the active sites of other thioredoxin-like oxidoreductases. A general acid/base catalytic mechanism has been proposed for E. coli thioredoxin on the basis of pKa values measured for the isolated protein (26). In the final step of the mechanism, the deprotonated carboxyl group of Asp26 (which is the structural counterpart of Asp455 in cDsbD) is proposed to abstract a proton from the thiol of Cys35, either directly or via a bridging water molecule (27), and the resulting thiolate attacks the mixed disulfide to form the reduced substrate and oxidized thioredoxin. A similar role for the analogous Asp/Glu is suggested in other thiol:disulfide oxidoreductases (26). The pKa value of 6.5 measured for Asp455 in isolated cDsbD suggests it could play a similar role. However, the elevated pKa value of 8.5 for Asp455 in the nDsbD-cDsbD complex makes this role unlikely. Other acidic groups in proximity to the thiol group of Cys464, such as Glu468, may play this role in cDsbD. An alternative mechanism for thioredoxin has been proposed in which the thiolate group in the substrate protein abstracts the proton from Cys35 (28, 29); this mechanism would also depend on the pKa value of the substrate thiol in the complex. Our results highlight the importance of considering the effects of protein-protein interactions in physiological complexes on the properties of active-site residues rather than simply considering protein domains in isolation.
Supplementary Material
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Experimental Procedures, Figs. S1 and S2, Tables S1 and S2, and additional references.
Author's Choice—Final version full access.
Footnotes
The abbreviations used are: BisTris, 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol; DDM, n-dodecyl β-d-maltoside; HRP, horseradish peroxidase; HSQC, heteronuclear single quantum coherence.
References
- 1.Kadokura, H., Katzen, F., and Beckwith, J. (2003) Annu. Rev. Biochem. 72111 -135 [DOI] [PubMed] [Google Scholar]
- 2.Ito, K., and Inaba, K. (2008) Curr. Opin. Struct. Biol. 18450 -458 [DOI] [PubMed] [Google Scholar]
- 3.Katzen, F., and Beckwith, J. (2000) Cell 103769 -779 [DOI] [PubMed] [Google Scholar]
- 4.Gordon, E. H., Page, M. D., Willis, A. C., and Ferguson, S. J. (2000) Mol. Microbiol. 351360 -1374 [DOI] [PubMed] [Google Scholar]
- 5.Collet, J. F., Riemer, J., Bader, M. W., and Bardwell, J. C. (2002) J. Biol. Chem. 27726886 -26892 [DOI] [PubMed] [Google Scholar]
- 6.Crooke, H., and Cole, J. (1995) Mol. Microbiol. 151139 -1150 [DOI] [PubMed] [Google Scholar]
- 7.Hodson, C. T., Lewin, A., Hederstedt, L., and Le Brun, N. E. (2008) J. Bacteriol. 1904697 -4705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goulding, C. W., Sawaya, M. R., Parseghian, A., Lim, V., Eisenberg, D., and Missiakas, D. (2002) Biochemistry 416920 -6927 [DOI] [PubMed] [Google Scholar]
- 9.Haebel, P. W., Goldstone, D., Katzen, F., Beckwith, J., and Metcalf, P. (2002) EMBO J. 214774 -4784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stirnimann, C. U., Rozhkova, A., Grauschopf, U., Bockmann, R. A., Glockshuber, R., Capitani, G., and Grutter, M. G. (2006) J. Mol. Biol. 358829 -845 [DOI] [PubMed] [Google Scholar]
- 11.Kim, J. H., Kim, S. J., Jeong, D. G., Son, J. H., and Ryu, S. E. (2003) FEBS Lett. 543164 -169 [DOI] [PubMed] [Google Scholar]
- 12.Rozhkova, A., Stirnimann, C. U., Frei, P., Grauschopf, U., Brunisholz, R., Grutter, M. G., Capitani, G., and Glockshuber, R. (2004) EMBO J. 231709 -1719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stirnimann, C. U., Rozhkova, A., Grauschopf, U., Grutter, M. G., Glockshuber, R., and Capitani, G. (2005) Structure (Lond.) 13985 -993 [DOI] [PubMed] [Google Scholar]
- 14.Stirnimann, C. U., Grutter, M. G., Glockshuber, R., and Capitani, G. (2006) Cell. Mol. Life Sci. 631642 -1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mavridou, D. A., Stevens, J. M., Ferguson, S. J., and Redfield, C. (2007) J. Mol. Biol. 370643 -658 [DOI] [PubMed] [Google Scholar]
- 16.Katzen, F., and Beckwith, J. (2003) Proc. Natl. Acad. Sci. U. S. A. 10010471 -10476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ausubel, F. M., Brent, R., Kingston, R. E., Moore, D. D., Seidman, J. G., Smith, J. A., and Struhl, K. (2003) Current Protocols in Molecular Biology, pp.16.1 -16.8, John Wiley & Sons, Inc., New York18265320
- 18.Delaglio, F., Grzesiek, S., Vuister, G. W., Zhu, G., Pfeifer, J., and Bax, A. (1995) J. Biomol. NMR 6 277-293 [DOI] [PubMed] [Google Scholar]
- 19.Johnson, B. A. (2004) Methods Mol. Biol. 278313 -352 [DOI] [PubMed] [Google Scholar]
- 20.Shrager, R. I., Cohen, J. S., Heller, S. R., Sachs, D. H., and Schechter, A. N. (1972) Biochemistry 11 541-547 [DOI] [PubMed] [Google Scholar]
- 21.Hiniker, A., Vertommen, D., Bardwell, J. C., and Collet, J. F. (2006) J. Bacteriol. 1887317 -7320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chung, J., Chen, T., and Missiakas, D. (2000) Mol. Microbiol. 351099 -1109 [DOI] [PubMed] [Google Scholar]
- 23.Li, H., Robertson, A. D., and Jensen, J. H. (2005) Proteins 61704 -721 [DOI] [PubMed] [Google Scholar]
- 24.Chivers, P. T., Prehoda, K. E., Volkman, B. F., Kim, B. M., Markley, J. L., and Raines, R. T. (1997) Biochemistry 3614985 -14991 [DOI] [PubMed] [Google Scholar]
- 25.Jeng, M. F., and Dyson, H. J. (1996) Biochemistry 351 -6 [DOI] [PubMed] [Google Scholar]
- 26.Chivers, P. T., and Raines, R. T. (1997) Biochemistry 3615810 -15816 [DOI] [PubMed] [Google Scholar]
- 27.Menchise, V., Corbier, C., Didierjean, C., Saviano, M., Benedetti, E., Jacquot, J. P., and Aubry, A. (2001) Biochem. J. 35965 -75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jeng, M. F., Holmgren, A., and Dyson, H. J. (1995) Biochemistry 3410101 -10105 [DOI] [PubMed] [Google Scholar]
- 29.Carvalho, A. T. P., Swart, M., van Stralen, J. N. P., Fernandes, P. A., Ramos, M. J., and Bickelhaupt, F. M. (2008) J. Phys. Chem. B 1122511 -2523 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.