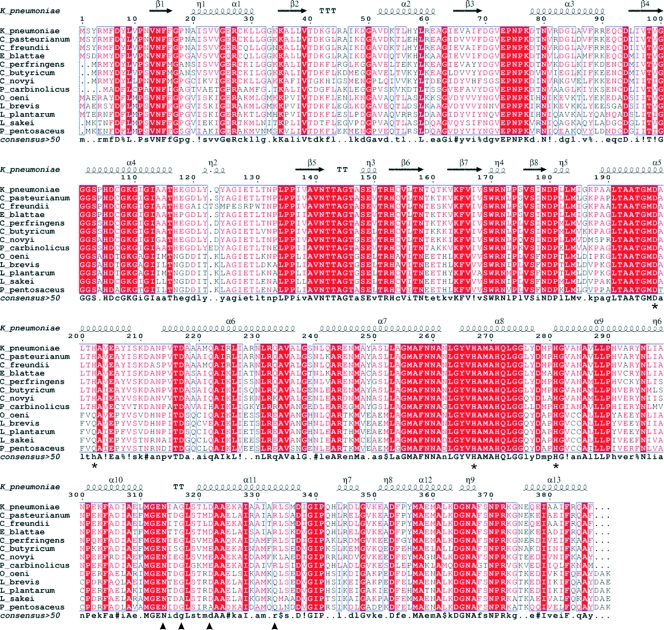
FIG. 1.
Sequence alignment of several members of the 1,3-PD dehydrogenase family. K_pneumoniae, Klebsiella pneumoniae; C_pasteurianum, Clostridium pasteurianum; C_freundii, Citrobacter freundii; E_blattae, Escherichia blattae; C_perfringens, Clostridium perfringens; C_butyricum, Clostridium butyricum; C_novyi, Clostridium novyi; P_carbinolicus, Pelobacter carbinolicus; O_oeni, Oenococcus oeni; L_brevis, Lactobacillus brevis; L_plantarum, Lactobacillus plantarum; L_sakei, Lactobacillus sakei; P_pentosaceus, Pediococcus pentosaceus. Highly conserved regions are boxed with similar residues are represented in red, and nonconserved residues are indicated in black. Within these boxes, invariant residues are represented against a red background. The secondary structure of 1,3-PD dehydrogenase from K. pneumoniae as derived from the three-dimensional data is represented in the upper part of the alignment. Residues involved in the binding of Fe atom are marked with an asterisk. Residues involved in decamer stabilization by dimer interactions are marked with a triangle. Alignment was prepared with Multalin (10) and the ENDscrip server (17).