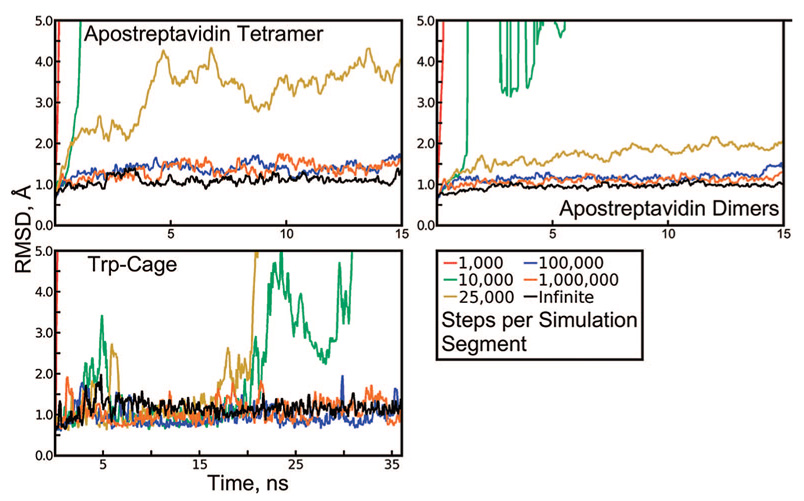
Figure 2.
Backbone rmsd of apostreptavidin and Trp-Cage, revealing artifacts in Langevin dynamics. Each protein was simulated in explicit solvent at 298 K using a Langevin thermostat with a collision frequency of 3 ps−1 and simulation segments with lengths given in the figure legend. The same random seed was used to reinitialize the pseudorandom number generator (PRNG) at the beginning of every segment, except for the “infinite” case, in which segments of 100 000 steps were initiated with different PRNG seeds every time.