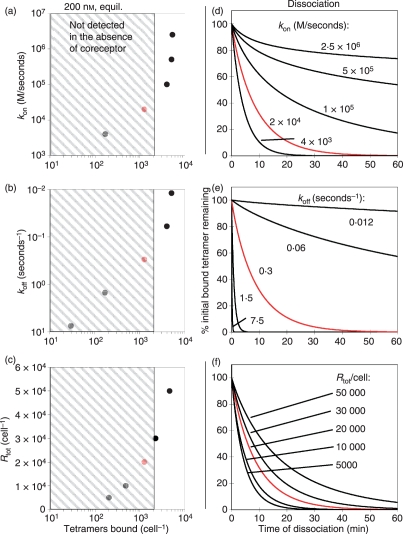
Figure 2.
Simulated effect of T-cell receptor : peptide–major histocompatibility complex (TCR:pepMHC) association kinetics, dissociation kinetics, or TCR surface levels on pepMHC tetramer binding. Using a simplified multivalent binding model, simulated values and curves were generated to predict (a–c) number of pepMHC tetramers bound per T cell or (d–f) dissociation rates of bound tetramers from a T cell. The varied TCR parameters included the kon (a,d), the koff (b,e), and the Rtot [total number of TCRs per T cell (c,f)]. In each panel, the fixed parameters correspond to the 2C TCR binding to SIY/Kb (kon = 20 000/m/second, koff∼0·3/second, Rtot∼20 000 per cell) and the simulated result for 2C TCR is shown in red. In panels (a–c) the equilibrium tetramer staining was simulated using 200 nm pepMHC tetramer, and the grey hatched boxes encompass the region below 2000 molecules per cell, a ‘threshold’ below which there may be no binding detectable by flow cytometry (i.e. detected as ‘no staining’ above the contol). Panels (d–f) show simulated tetramer dissociation rates predicted by varying the indicated parameters. The potential contribution of CD8 to binding is not assessed.