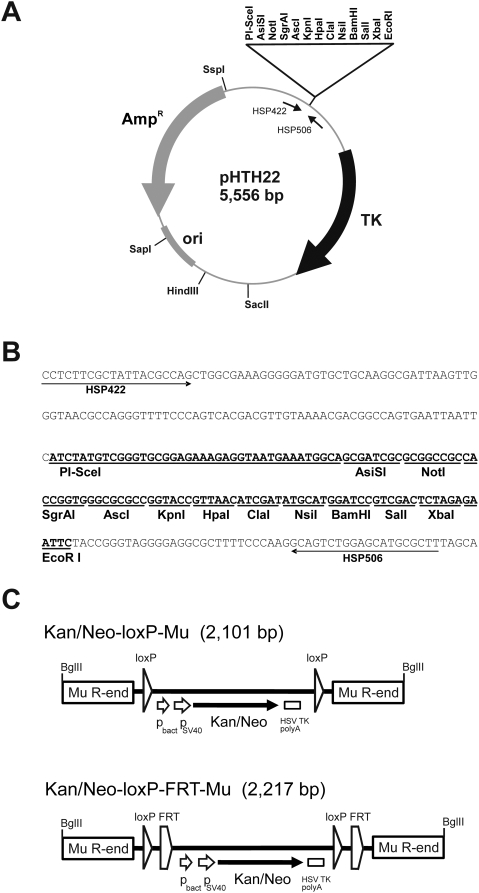
Figure 1. Details of the genetic tools used.
(A) Structure and characteristic elements of the multicopy cloning vector pHTH22 with unique restriction sites indicated. The gene region encoding HSV thymidine kinase (TK) under the control of mouse phosphoglycerate kinase promoter and terminator is shown with a black curved arrow. The vector portion of the plasmid including the gene for ampicillin resistance (AmpR) and pUC19 origin of replication (ori) are shown with gray symbols (curved arrow and rectangle). (B) The nucleotide sequence of the polylinker and its flanking regions in pHTH22. The polylinker is shown in boldface with enzyme recognition sites underlined. The arrows indicate the binding sites of primers that were used for the confirmatory sequencing of genomic inserts' ends. (C) Structures of the Kan/Neo-loxP-Mu and Kan/Neo-loxP-FRT-Mu transposons. The transposons contain bacterial (pbact) and eukaryotic (pSV40) promoters (short arrows), a marker gene (Kan/Neo) conferring resistance to kanamycin in bacteria and G418 in eukaryotes (black arrow), and the HSV thymidine kinase polyadenylation (HSV TK polyA) signal (small rectangle). The loxP sites are indicated by triangles and FRT sites by pentagons. The rectangles in the transposon ends indicate 50 bp of Mu R-end DNA sequences in inverted orientation relative to each other. For the sake of clarity, the features are not in scale. The BglII sites in the ends are used to excise the transposons from their carrier plasmids. The sequences of the transposon-containing plasmids pHTH19 (Kan/Neo-loxP-Mu) and pHTH24 (Kan/Neo-loxP-FRT-Mu) are available upon request.