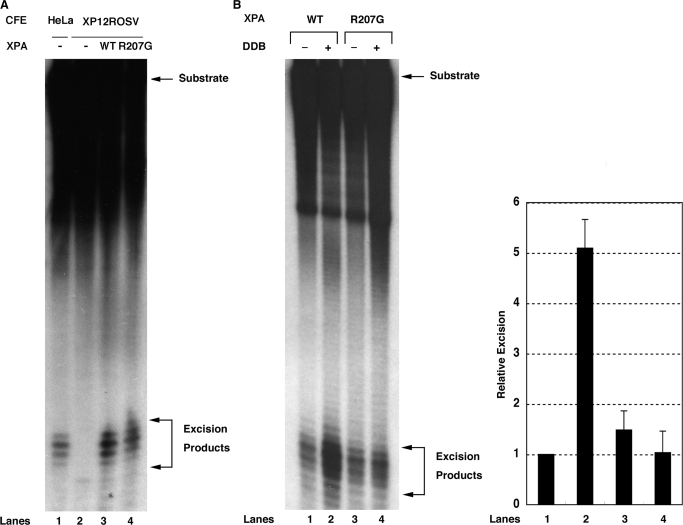
Figure 4.
Effect of R207G mutation on XPA activity in NER reaction in vitro. (A) Three femtomoles of internally-labeled 136-bp substrates containing a 6-4PP were incubated with 50 µg of XP-A CFEs in the absence (lane 2) or presence of 60 ng of wild-type (lane 3) or R207G mutant XPA (lane 4) for 45 min. As a control, the excision reaction was also conducted with 50 µg of HeLa CFEs (lane 1). DNAs were extracted, separated on an 8% sequencing gel and detected by autoradiography. The signals were quantified by a Fuji Bas 2000 Bio-imaging analyzer and each excision efficiency was determined as 0.66% (lane 1), 0.017% (lane 2), 2.2% (lane 3) and 2.9% (lane 4). (B) Six femtomoles of internally-labeled 136-bp substrates containing a CPD were incubated with 60 ng of wild-type (lanes 1 and 2) or R207G mutant XPA (lanes 3 and 4) and other core NER factors (150 ng of RPA, 20 ng of XPC-RAD23B, 150 ng of TFIIH, 10 ng of XPG and 20 ng of XPF-ERCC1) in the presence (lanes 2 and 4) or absence (lanes 1 and 3) of DDB heterodimer (50 ng) for 4 h. DNAs were analyzed as described in (A) and each excision efficiency was 0.98% (lane 1), 5.6% (lane 2), 1.2% (lane 3) and 0.74% (lane 4). A right panel shows quantitative data from triplicate experiments and percent excision was expressed as relative excision to control reaction (wild-type XPA, no DDB). Error bars indicate the SD.