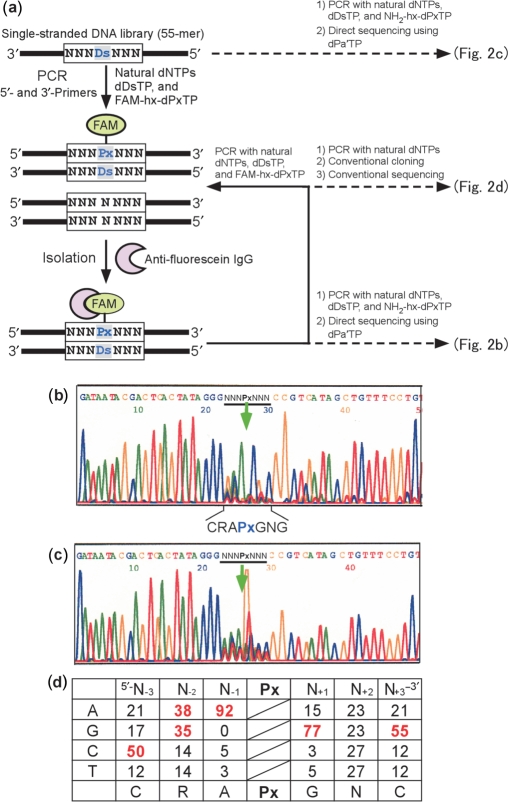
Figure 2.
Overview of in vitro selection using a Ds-containing DNA library. (a) Scheme for in vitro selection using the Ds-containing DNA library, by FAM-hx-Px incorporation into PCR products and isolation with an anti-fluorescein antibody. A chemically synthesized, single-stranded DNA library containing an NNNDsNNN sequence (55-mer) was amplified by 10 cycles of PCR, in the presence of natural dNTPs, dDsTP and FAM-hx-dPxTP, with DeepVent DNA Pol. After selection of the PCR products containing FAM-hx-Px by binding with an anti-fluorescein antibody, the isolated DNA fragments were used as a template for the next round of PCR amplification and selection. For direct sequencing of the library after five rounds of selection, the isolated DNA fragments were amplified in the presence of NH2-hx-dPxTP, instead of FAM-hx-dPxTP (Figure 3a), by eight cycles of PCR. Direct sequencing was performed in the presence of 2 μM dPa′TP, using a BigDye terminator v1.1 Cycle Sequencing kit. For the sequencing of clones after five rounds of selection, the library was amplified in the presence of only the natural dNTPs with Taq DNA polymerase, and the PCR products were used for TOPO TA cloning. (b) Sequencing of the DNA library after five rounds of selection. (c) Sequencing of the initial library. The arrow indicates the unnatural base position. (d) Probability (%) of occurrence at each position of the selected 66-clone sequences (Supplementary Figure 1). Bases with an occurrence rate of ≥35% among the clones are colored red.