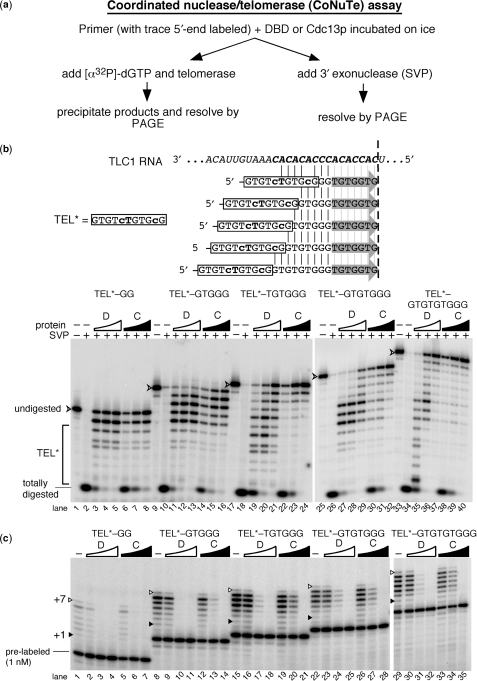
Figure 5.
A coordinated assay reveals that Cdc13p and its DBD inhibit telomerase similarly via distinct DNA interactions. (a) CoNuTe assay scheme used to generate data in b and c. (b) SVP assay shows that Cdc13(DBD) (D) preferentially protects TEL* even in the context of long 3′ telomeric appendages, while Cdc13p (C) protects the 3′ region of substrates with ⩾6-nt tails. Oligonucleotides tested are shown: TEL* region is boxed and vertical lines indicate complementarity to TLC1 RNA shown above, while gray arrows indicate the 7 TLC1-endoded nucleotides predicted to be added to each by telomerase (results for which are shown in c). SVP results are shown below. V-shaped arrowhead indicates completely undigested 5′-end labeled primer. TEL*, region of gel with SVP products 11 nucleotides or shorter. Arrowhead indicates full-length 5′-end labeled oligonucleotide. Totally digested, DNA not protected by binding to DBD or Cdc13p; serves as an indicator for fraction complex formation. Concentrations of Cdc13(DBD) were 0.15 µM, 0.3 µM and 0.6 µM and for Cdc13p were 0.3 µM, 0.6 µM and 1.2 µM. (c) Telomerase is consistently inhibited by the DBD and Cdc13p bound to TEL*-based oligonucleotides containing 2–9-nt appendages. Pre-labeled 5′-end-labeled primers, included in the reaction for the SVP experiment shown in (b), are indicated for the shortest primer and serves as a recovery and loading control for telomerase activity assays. The +1 and +7 telomerase products are indicated as in Figures 1 and 2 (gray arrows in b). See legend b for concentrations of DBD and Cdc13p.