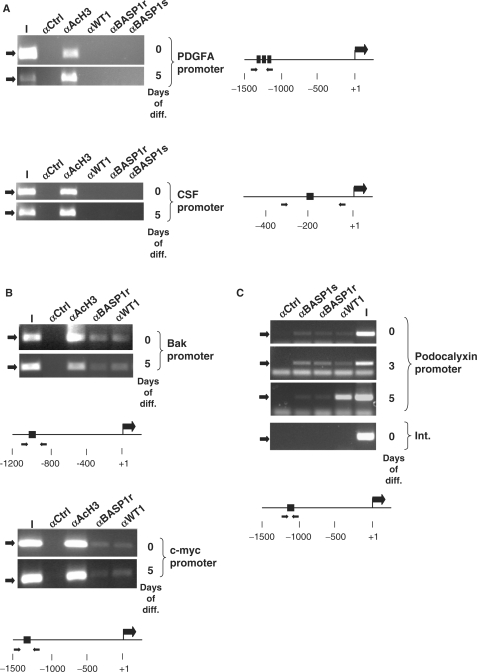
Figure 3.
Promoter occupancy by WT1 and BASP1 in differentiating MPC5 cells. (A) ChIP was performed with undifferentiated MPC5 cells (0) or MPC5 cells that had differentiated for 5 days (5). The antibodies used in the ChIP were rabbit anti-GAL4 antibodies (αCtrl), anti-acetylated histone H3 (αAcH3), rabbit anti-WT1 antibodies (αWT1), rabbit anti-BASP1 antibodies (αBASP1r) and sheep anti-BASP1 antibodies (αBASP1s). The resultant samples were subject to amplification by PCR using primers directed to the mouse PDGFA promoter (top) or CSF1 promoter (bottom). I is 1/50 of the input chromatin in each immunoprecipitation. A diagram of each promoter region is shown at right, with the positions of the WT1 DNA-binding sites and forward/reverse primer regions indicated. (B) ChIP was performed as in part A except that the Bak (top) and c-myc (bottom) promoters were analysed. (C) As in (A) except that the analysis was performed with undifferentiated MPC5 cells (0) and cells that had been allowed to differentiate for 3 (3) and 5 (5) days. The bottom panel is amplification of an internal region of the podocalyxin gene (Int.) from ChIP analysis of undifferentiated MPC5 cells.