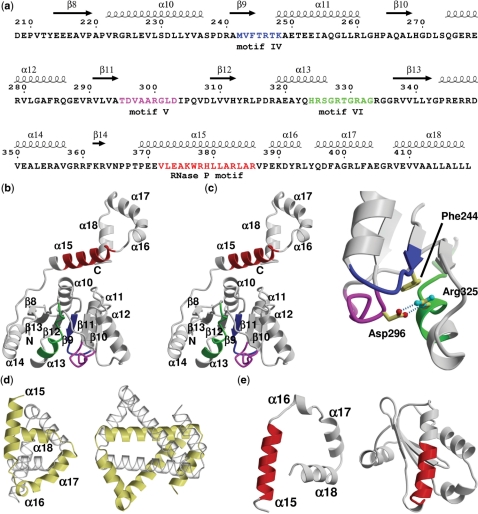
Figure 1.
Hera structure sequence relationship and architecture of the Hera_208–419 monomer. (a) Sequence of Hera_208–419 with secondary structure elements indicated on the top. The numbering of sequence and secondary structure elements corresponds to the full-length Hera. Conserved helicase motifs IV–VI are colored blue, magenta and green, respectively. The putative RNase P motif is colored in red. (b) Stereo ribbon diagram of the Hera_208–419 monomer. The secondary structure elements, helicase and RNase P motifs from (a) are indicated. (c) Close-up showing the interactions of helicase motifs IV–VI. Asp296 of motif V connects to Arg325 motif VI via two hydrogen bonds (dashed lines). Arg325 stacks on Phe244 of motif IV. The view is rotated by 180° around the y-axis compared to (b). (d) Comparison of the left-handed super-helices in Hera_208–419 (left) and the hypothetical H. pylori protein HP242 (right). The corresponding monomers are colored identically in yellow and transparent grey. (e) The RNase P motifs in Hera (left) and in the protein component of the T. maritima RNase P (PDB-ID 1nz0; right) are predominantly α-helical but located in a very different structural context. The structures are shown with their RNase P motifs aligned (red).