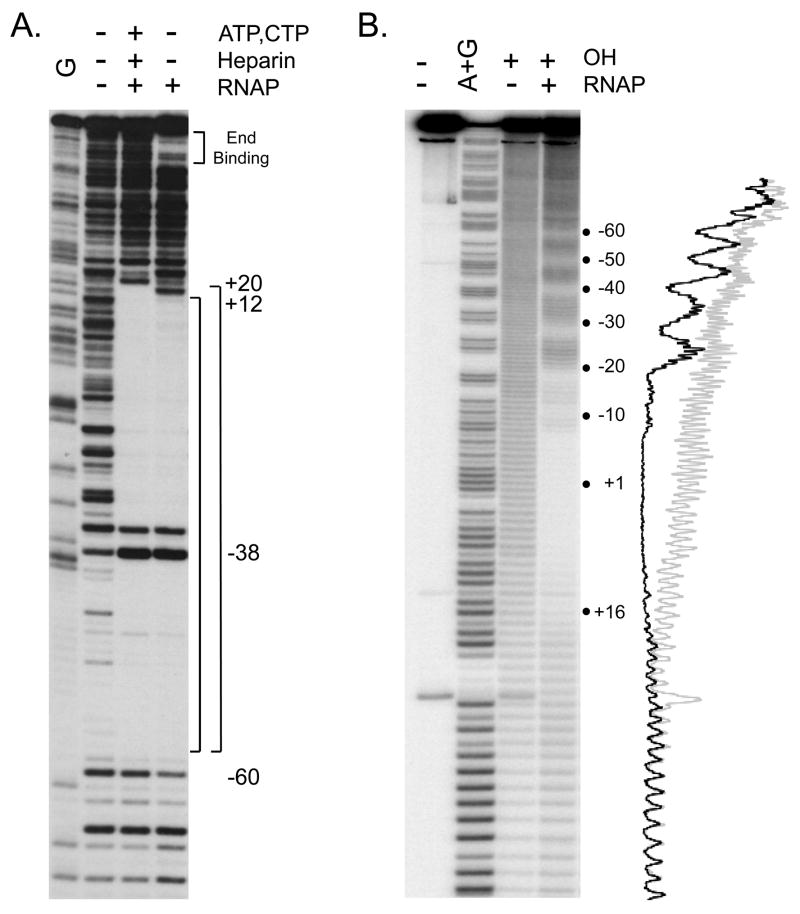
Fig. 4.
(A) DNAse I footprints of RNAP complexes formed with the E. coli rrnB P1 promoter. The promoter fragment (−88 to +50) was 32P-end labeled at the 3′ end of the template strand. Complexes were formed with 10 nM RNAP at 37°C in buffer containing 30 mM KCl, 10 mM Tris-Cl pH7.9, 10 mM MgCl2, 1 mM DTT and 100 μg/ml BSA, and were digested with DNase I (0.6 ug/ml) for 30 sec. The sample in lane 3 also contained the two initiating nucleotides ATP (500 μM) and CTP (50μM), resulting in formation of a heparin stable complex, RPAC [32, 58], and heparin was added to this sample prior to DNAse I digestion. The upstream boundary of protection against DNaseI digestion (~−60) is the same in lanes 3,4 and the downstream boundary differs (+20 for RPAC, lane 3; ~+12 for the complex formed in the absence of ATP and CTP, lane 4). Position of DNaseI enhancement is noted at −38. Binding of RNAP to the end of the promoter fragment is indicated in lane 4, but is not seen in the presence of heparin (lane 3). The sequence marker (G ladder; lane 1) was prepared by dimethyl sulfate modification, followed by strand cleavage with piperidine [80]. (B) Hydroxyl radical footprint of an RNAP-lacUV5 promoter complex. The lacUV5 fragment (−130 to +40) was 32P- labeled at the 3′ end of the non-template strand, and was incubated with 15 nM RNAP at 37°C in a buffer containing 110 mM KCl, 10 mM Tris-Cl pH 8.0, 10 mM MgCl2, 1 mM DTT, followed by hydroxyl radical cleavage as described in text. The RNAP-promoter complex was then separated from free DNA by retention on a nitrocellulose filter and eluted from the filter (see text). Lane 1: promoter fragment, no digestion; lane 2: A+G marker prepared as described in text; lane 3: promoter fragment digested with hydroxyl radicals in absence of RNAP; lane 4: RNAP-promoter complex digested with hydroxyl radicals. Traces of gel lanes 3 (light gray) and 4 (black) were generated as described in text.