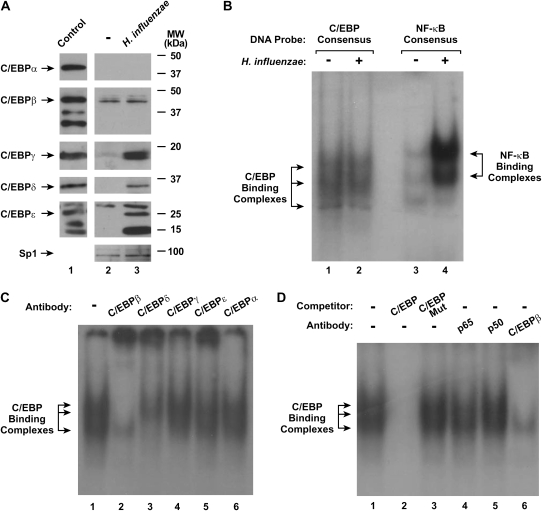
Figure 4.
H. influenzae affects nuclear expression of C/EBP family members. (A) C/EBP family and Sp1 (to verify equivalent isolation and loading) nuclear protein levels were assessed using immunoblot analysis of nuclear protein extracts from hTBE cell monolayers that were incubated without or with H. influenzae for 4 hours. Control samples were rat liver nuclear extract (Active Motif, Carlsbad, CA) for C/EBPα, THP-1 cell nuclear extract for C/EBPβ and C/EBPδ, purified C/EBPγ amino acids 39–147 linked to a His-tag (US Biological, Swampscott, MA), and HL-60 nuclear extract (Abcam, Cambridge, MA) for C/EBPɛ. The positions of C/EBP family members and control Sp1 are indicated by arrows. (B) Transcription factor binding to C/EBP and NF-κB consensus sequences that were similar in nucleotide number was assessed using EMSA with nuclear extracts from hTBE cell monolayers that were incubated without or with H. influenzae for 4 hours. The position of C/EBP- and NF-κB–binding complexes are indicated by arrows. (C) Transcription factor binding to a C/EBP consensus sequence was assessed using EMSA with nuclear extracts from hTBE cells that were incubated with H. influenzae for 4 hours. Supershift analysis was performed by addition of antibodies against C/EBPβ, C/EBPδ, C/EBPγ, C/EBPɛ, or C/EBPα. (D) Transcription factor binding to a C/EBP consensus sequence was assessed using EMSA with nuclear extracts from hTBE cells that were incubated with H. influenzae for 4 hours. Specificity of protein binding was assessed by competition with unlabeled oligonucleotides containing the C/EBP consensus sequence without (C/EBP) or with (C/EBP Mut) mutation. Supershift analysis was performed by addition of antibodies against NF-κB (p65 or p50) or C/EBPβ. Results in all panels are representative of three to five experiments.