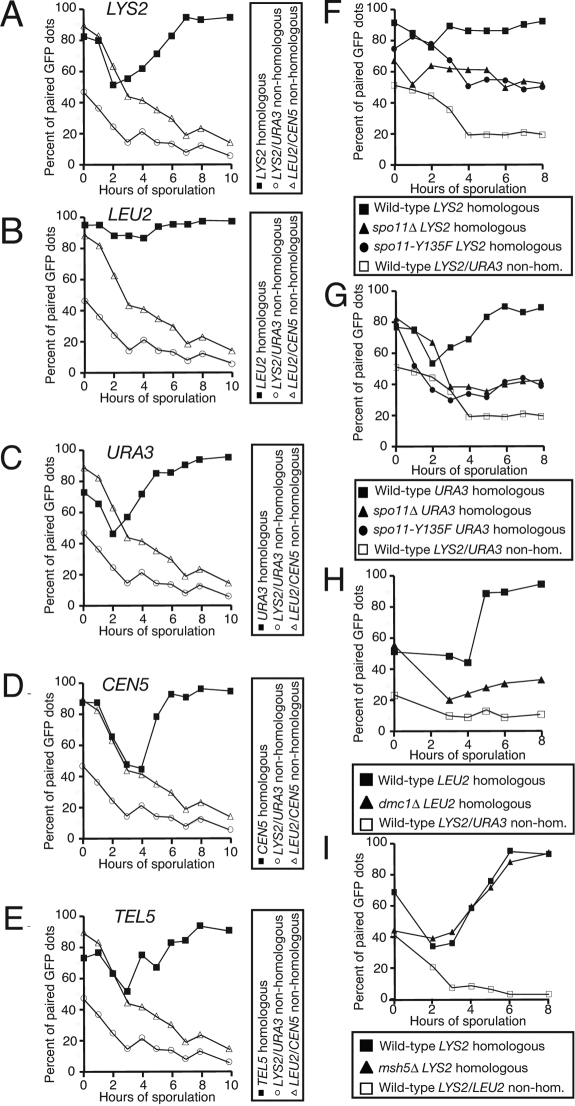
Figure 1.
An assay to monitor pairing in live cells. (A) Wild-type cells with homologous LYS2 dots (A9828, ■), wild-type cells with nonhomologous LYS2/URA3 dots (A9829, ○), and wild-type cells with nonhomologous LEU2/CEN5 dots (A16360, ▵), all deleted for NDT80, were introduced into sporulation medium. At the indicated times, samples were taken and assayed for pairing live on aliquots that are taken at the indicated times as cells progress through prophase. Paired GFP signals are not distinguishable, and only a single GFP dot is visible. Nonhomologous arrays are included in each experiment as a control for clustering of the tet operators. Note that the same nonhomologous dot controls are used in A–E as the data from these panels were generated from the same experiment. (B) Wild-type cells with homologous LEU2 dots (A5111, ■), wild-type cells with nonhomologous LYS2/URA3 dots (A9829, ○), and wild-type cells with nonhomologous LEU2/CEN5 dots (A16360, ▵), all deleted for NDT80, were assayed for pairing as described in A. (C) Wild-type cells with homologous URA3 dots (A6946, ■), wild-type cells with nonhomologous LYS2/URA3 dots (A9829, ○), and wild-type cells with nonhomologous LEU2/CEN5 dots (A16360, ▵), all deleted for NDT80, were assayed for pairing as described in A. (D) Wild-type cells with homologous CEN5 dots (A16362, ■), wild-type cells with nonhomologous LYS2/URA3 dots (A9829, ○), and wild-type cells with nonhomologous LEU2/CEN5 dots (A16360, ▵), all deleted for NDT80, were assayed for pairing as described in A. (E) Wild-type cells with homologous TEL5 dots (A16366, ■), wild-type cells with nonhomologous LYS2/URA3 dots (A9829, ○) and wild-type cells with nonhomologous LEU2/CEN5 dots (A16360, ▵), all deleted for NDT80, were assayed for pairing as described in A. (F) Wild-type cells with homologous LYS2 dots (A9828, ■), spo11Δ cells with homologous LYS2 dots (A11469, ▴), spo11-Y135F cells with homologous LYS2 dots (A12095, ·), and wild-type cells with nonhomologous LYS2/URA3 dots (A9829, □), all deleted for NDT80, were assayed for pairing as described in Figure (A). Note that the same nonhomologous dot control strain is used in F and G as the data from these panels were generated from the same experiment. (G) Wild-type cells with homologous URA3 dots (A6946, ■), spo11Δ cells with homologous URA3 dots (A7803, ▴), spo11-Y135F cells with homologous URA3 dots (A9115, ·), and wild-type cells with nonhomologous LYS2/URA3 dots (A9829, □), all deleted for NDT80, were assayed for pairing as described in A. (H) Wild-type cells with homologous LEU2 dots (A5111, ■), dmc1Δ cells with homologous LEU2 dots (A6917, ▴), and wild-type cells with nonhomologous LEU2/LYS2 dots (A11474, □), all deleted for NDT80, were assayed for pairing as described in A. (I) Wild-type cells with homologous LYS2 dots (A9828, ■), msh5Δ cells with homologous LYS2 dots (A16290, ▴) and wild-type cells with nonhomologous URA3/LYS2 dots (A9829, □), all deleted for NDT80, were assayed for pairing as described in A.