Figure 4. Three-Dimensional Reconstruction of Hsc82:AMPPNP.
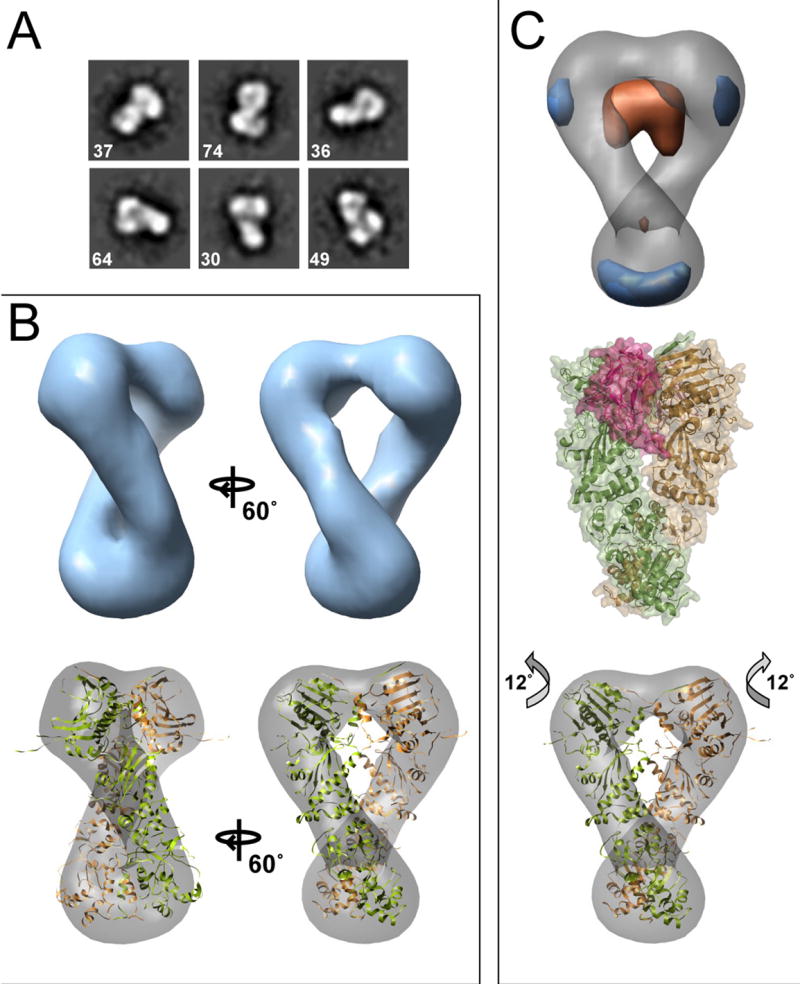
Reference-free class averages are shown in (A). In (B) the final Hsc82:AMPPNP reconstruction is shown along with the Hsp82:AMPPNP:p23 crystal structure docked into the map. In (C) a difference map (experimental - calculated) is shown aligned to the yeast Hsc82:AMPPNP reconstruction. In orange is density present in the crystal structure but missing in the EM volume, and blue is additional EM map density. For comparison, the yeast Hsp82:AMPPNP:p23 complex is shown (C, middle panel). Two p23 molecules (raspberry) flank the NTD dimerization interface. An improved, rigid-body docking of the domains into the EM map (C, lower panel) highlights an alternate closed conformation with a 12° rotation of the NTD.
