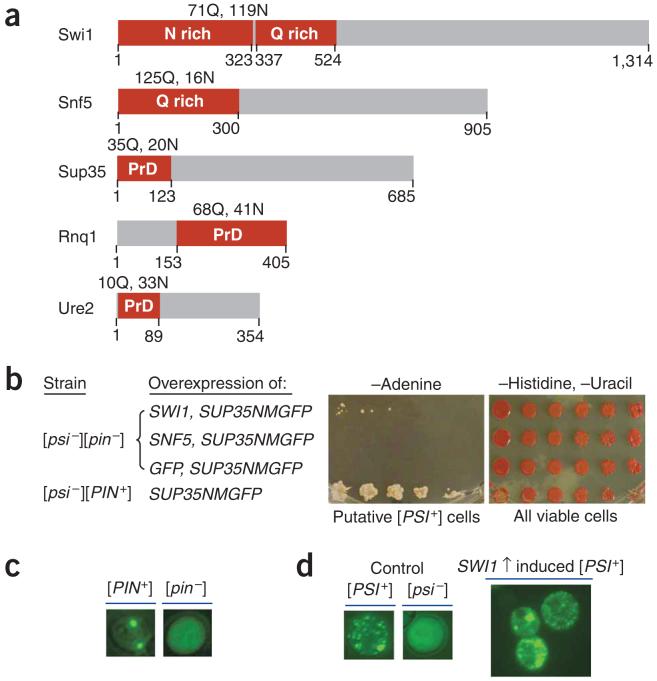
Figure 1.
Overexpression of SWI1 but not SNF5 functions as Pin+. (a) Diagrams of Swi1 and Snf5 with their glutamine (Q) and asparagine (N) rich regions shown (red boxes). The prion domains (PrDs) of Sup35, Rnq1 and Ure2 are also indicated (red boxes). (b) Overexpression of SWI1 can substitute for [PIN+] to promote [PSI+] formation. We grew 74D-694 cells [(psi-][pin-]) containing pRS313CUP1NMGFP and p426GPDSWI1, pRS313CUP1NMGFP and p426GPDSNF5, or pRS313CUP1NMGFP and p426GPDGFP to early-log phase and induced them for NMGFP expression by addition of CuSO4 to 100 μM. After 24-h induction, cells were spotted onto the indicated plates with a fivefold serial dilution. Pictures were taken after 5-d incubation for cells on synthetic complete medium lacking histidine and uracil (-Histidine, -Uracil) or after 9 d for cells on medium lacking adenine (-Adenine). (c) Fluorescence microscopy assay of Rnq1 conformational status. Before conducting experiments shown in panel b, we confirmed the Rnq1 conformational status by transforming with p416CUP1RNQ1GFP, inducing for 4 h with 100 μM CuSO4 and analyzing by fluorescence microscopy. (d) Sup35 is aggregated in Ade+ cells obtained by SWI1 and SUP35NMGFP co-overexpression. After eliminating p426GPDSWI1 and pRS313CUP1NMGFP, we transformed Ade+ isolates from panel b with p416CUP1NMGFP and analyzed them by fluorescence microscopy to confirm their [PSI+] status. Shown here are fluorescence patterns of a representative [PSI+] isolate and isogenic [PSI+] and [psi-] controls.