Abstract
Neuroblasts migrate from the subventricular zone along the rostral migratory stream (RMS) to the olfactory bulb (OB). While the migration occurs by movement over other cells, the molecular mechanisms are poorly understood. We have found that ADAM2 (a disintegrin and metalloprotease 2) is expressed in migrating RMS neuroblasts and functions in their migration. The brains from ADAM2 knockout (KO) mice showed a smaller OB than that seen in wild-type (WT) mice at postnatal day 0. In addition, the RMS in ADAM2 KO mice appeared thinner and less voluminous in its rostral part and thicker in its caudal part. Estimates of migration in vivo using bromodeoxyuridine labeling revealed that neuroblasts from KO mice show a decreased migration rate compared with those from WT mice. Direct assays of migration by imaging living slices also showed a decreased migration speed and loss of directionality in the KO mice. This phenotype was similar to that seen in RMS containing slices from WT mice exposed to a peptide that mimicked the disintegrin loop of ADAM2. Finally, RMS explants from KO or WT mice that were cultured in Matrigel also revealed striking differences. The cells migrating out of explants from WT mice showed robust cell—cell interactions. In contrast, fewer cells migrated out of explants from ADAM2 KO mice, and those that did were largely dispersed and their migration inhibited. These experiments suggest that ADAM2 contributes to RMS migration, possibly through cell—cell interactions that mediate the rapid migration of the neuroblasts to their endpoint.
Keywords: ADAM2, chain migration, integrin, mouse, neurophilic contact, slice culture
Introduction
Identifying the molecules that mediate neuroblast migration is central to understanding molecular mechanisms of brain development and congenital defects that compromise brain organization and function. It also holds promise for developing therapeutic strategies for the repair of damaged brain regions due to trauma or neurodegenerative diseases. The rostral migratory stream (RMS), which begins at the anterior part of the subventricular zone (SVZa) and ends at the center of the olfactory bulb (OB), is an excellent system for studying neuroblast migration (Doetsch et al., 1999). Initially, RMS neuroblasts migrate tangentially, and then upon reaching the OB they migrate radially toward the periphery of the OB where they differentiate into mature neurons (Altman, 1969; Luskin, 1993). The tangential migration is not associated with either glial or axonal fibers (Kishi et al., 1990) and, instead, the leading processes of the neuroblasts overlap somata of other neuroblasts resulting in the formation of long chains of neuroblasts over which the cells appear to migrate both in vivo and in vitro (Rousselot et al., 1995; Lois et al., 1996; Wichterle et al., 1997). These migrations are based on cell—cell interactions and termed neurophilic migration (Rakic, 1985).
Several molecules have been implicated in RMS migration. While reelin, tenascin-R and prokineticin 2 appear to initiate detachment of neuroblasts from chains and facilitate their radial migration (Hack et al., 2002; Saghatelyan et al., 2004; Ng et al., 2005), the molecules that mediate the interactions that form chains are unclear. Recently, members of the ADAMs (a disintegrin and metalloprotease) family have been observed in the nervous system (Sagane et al., 1998, 2005; Karkkainen et al., 2000; Leighton et al., 2001; Yang et al., 2005; Takahashi et al., 2006), and may function in neuronal migration. An ADAM is a multidomain, membrane-spanning protein containing a metalloprotease domain and a disintegrin domain, that often has protease- and/or integrin-binding activity (Blobel et al., 1992; Wolfsberg et al., 1993). There are 22 ADAMs in humans (White, 2003; Huovila et al., 2005). ADAM2, also called fertilin β, is expressed in mammalian sperm and is reported to play an important role in sperm—egg adhesion (Myles et al., 1994; Cho et al., 1998). However, its presence and function in other tissues is not well studied, although it has been reported in the brain (Karkkainen et al., 2000). In the present study, we investigated the distribution of ADAM2 in mouse brain using immunohistochemistry and in situ hybridization, and found that ADAM2 is expressed continually, from a late embryonic stage to adult, in migrating neuroblasts in the RMS. We also show that it contributes to the directed migration of neuroblasts in the RMS based on several different observations, including: direct imaging of the migrating cells in slices from ADAM2 knockout (KO) mice; perturbation of ADAM2 function with a specific peptide in explants from normal mice; and in vivo measurements of migration.
Materials and methods
ADAM2 KO mice
The ADAM2 KO mice have been described previously (Cho et al., 1998). ADAM2 exon 14, which contains the disintegrin domain active site (loop), was disrupted by homologous recombination. They were backcrossed 10 generations into a background of the ICR strain of mice, and the mutated gene was maintained in ICR strains. Homozygous mice were identified by polymerase chain reaction (PCR) of genomic DNA. Three oligonucleotide primers were used with the following sequences:
NeoB (KO primer): 5′-TTCGCAGCGCATCGCCTTCTATCG-3′; FBAct site (WT primer): 5′-TGCAGGCTTGCCCAAGATGAGTGTG-3′; 1.35B: 5′-AGACCGGGCAACCACTTCTAACGG-3′.
The PCR was done under the following conditions: denature 1 min @94 °C; anneal 2 min @62 °C; and extend 3 min @72 °C. The reaction was repeated for 40 cycles, and the reaction products were run on a 1.2% agarose gel. Primers 1 and 3 amplify a 1.8-kB product from the targeted ADAM2 allele. Primers 2 and 3 amplify a 2.1-kB band from the wild-type (WT) ADAM2 allele. PCR of heterozygous animals yielded both amplification products. ADAM2 KO and age-matched WT littermates were derived from heterozygous parents, and used for analysis.
Immunohistochemistry
ICR strain mice were purchased from Hilltop Laboratory Animals (Scottdale, PA, USA) and Charles River Laboratories (Wilmington, MA, USA), and housed using a 12 h light : dark cycle. For immunostaining, the mice were anesthetized with halothane. To obtain the brain of fetuses, deep anesthesia was used on the pregnant mice, and the abdominal cavity was opened to remove the fetuses. The mother’s diaphragm was cut to ensure that she had been killed. The embryos still under the crossover anesthesia were perfused through the aorta with fixatives consisting of either 4% paraformaldehyde/0.1 M phosphate buffer or acid ethanol (5% acetic acid in ethanol) or ice-cooled methanol. The postnates were anesthetized with halothane and perfused through the aorta with the above fixatives. The dissected brains were immersed in 25% sucrose/phosphate-buffered saline (PBS), frozen in liquid nitrogen and embedded in Tissue-Tek OCT compound (Miles, IN, USA). Sections (20 μm) of brains were cut using a cryostat, and mounted on silane-coated slides. For immunoperoxidase staining, sections were treated with primary antibodies for 42 h at 4 °C and then incubated with peroxidase-conjugated secondary antibodies for 2 h at room temperature. The immune complexes on the sections were detected using a peroxidase substrate consisting of diaminobenzidine-tetrahydrochloride. To demonstrate the localization of ADAM2 protein when using clone 9D2.2 against ADAM2, we performed heat-induced antigen retrieval by boiling the tissue sections prior to primary antibody incubation (Yamashita & Okada, 2005). After rinsing to remove unbound primary antibodies, the sections were incubated with Alexa-Fluor 488-conjugated anti-mouse IgG (Molecular Probes, Eugene, OR, USA) and examined in a laser-scanning confocal microscope FV300 (Olympus, Melville, NY, USA). The University of Virginia Animal Care and Use Committee approved all procedures.
Antibodies
A rabbit polyclonal antibody against ADAM2 disintegrin loop (Bigler et al., 2000), a mouse monoclonal antibody clone 9D2.2 against recombinant ADAM2 (Chemicon International, Temecula, CA, USA) and a polyclonal antibody against cytoplasmic domain of ADAM2 (Chemicon, AB19030) were used. Clone 2-2B for polysialic acid (PSA; Rougon et al., 1986) was purchased from Chemicon and used as a marker for migrating neuroblasts in the RMS (Rousselot et al., 1995). Anti-glial fibrillary acidic protein (GFAP, clone GA5, Millipore, Billerica, MA, USA; Debus et al., 1983) and anti-tenascin-C (clone MTn-12, Sigma-Aldrich, St Louis, MO, USA; Aufderheide & Ekblom, 1988) antibodies were used as glial markers. Anti-neurofilament antibody (clone 3A10, Developmental Studies Hybridoma Bank, University of Iowa, Department of Biological Sciences, Iowa City, IA, USA; Dodd et al., 1988) was used as a marker for mature neurons. Anti-tyrosine hydroxylase (TH) monoclonal antibody (clone 2/40/15) and anti-calbindin polyclonal antibody (CB38) were purchased from Chemicon and Swant (Bellinzona, Switzerland), to visualize periglomerular dopaminergic and calbindin-positive cells and interneurons in the granular cell layer (GCL; Bastianelli & Pochet, 1995), respectively. The specificities of the polyclonal antibodies were verified by absorbing each primary antibody with 1–2 mM of corresponding antigen—peptide solution overnight at 4 °C prior to application to brain sections, which was then followed by the diaminobenzidine reaction. Other control experiments included omission of primary antibodies.
In situ hybridization
Tissue sections from paraformaldehyde-fixed brains were prepared as described above. The sections were air dried for 5 min prior to hybridization with oligonucleotide probes for ADAM2. After rinsing in PBS, depurination was performed for 20 min with 0.2 M HCl at room temperature, and the tissues were treated with proteinase K for 15 min at 37 °C. After postfixation with 4% paraformaldehyde in PBS (5 min), the sections were immersed twice for 30 min in 2 mg/mL glycine in PBS. The sections were dehydrated with a series of solutions of increasing ethanol concentration and chloroform, and then dried by air. An ‘antisense’ oligonucleotide probe (5′-TGA CAT CAC ACT CAT CTT GGG CAA GCC TGC AAA CCT CCC C-3′) complementary to the sequence encoding a part of disintegrin domain of mouse ADAM2 was employed for hybridization experiments. ‘Sense’ strand probes with target sequences complementary to those of the antisense probes were used as a control for non-specific hybridization. The oligonucleotides were chemically synthesized and high-performance liquid chromatography (HPLC)-purified by Integrated DNA Technologies (Coralville, IA, USA). The oligoprobes (0.1 μmol) were labeled at the 3′ end with digoxigenin. Hybridization was performed at 37 °C for 12 h with 0.1 μg/mL digoxigeninoligonucleotide probe dissolved in the hybridization medium. After washing at 37 °C with 2 × standard saline citrate (SSC) for 30 min, 1 × SSC for 30 min and 0.5 × SSC for 30 min twice, the sections were incubated in blocking solution (Boehringer Mannheim, Mannheim, Germany) for 1 h. The sections were then treated for 2 h with alkaline phosphatase-labeled anti-digoxigenin antibody (Boehringer Mannheim) diluted (1 : 300) with the blocking solution. The alkaline phosphatase substrate 5-bromo-4-chloro-3-indolyl phosphate nitroblue tetrazolium chloride was used for color development, without counterstaining. All solutions used in this experiment were autoclaved in the presence of 0.1% diethylpyrocarbonate.
Preparation of brain slices
Slices were prepared from P10 mice using the protocol described elsewhere (Murase & Horwitz, 2002; Webb et al., 2002). For each individual experimental determination, a minimum of two slices from each brain was used, and the results confirmed by at least two additional determinations. Parasagittal slices that contained the entire RMS, which originated from the SVZa and ended at the center of the OB, were selected. The postnates were anesthetized with halothane and decapitated. The brains were placed into cell culture media (CCM)1 (HyClone Laboratories, Logan, UT, USA) medium at 4 °C. The brains were then transferred into CCM1 containing 10% heatinactivated horse serum (Sigma) and penicillin-streptomycin antibiotics (Sigma). They were then embedded in CCM1 medium containing 8% agarose (Sigma Type IX) and sliced into 200-μm sections using a vibratome. The slices were cultured on a Millicell-CM membrane (Millipore, Bedford, MA, USA) according to an original protocol (Stoppini et al., 1991).
Fluorescent labeling of the neuroblasts and time-lapse videomicrography
Small crystals of DiI (1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate, D-3911, Molecular Probes, Eugene, OR, USA) were placed onto the elbow of the RMS (RMSe; Pencea & Luskin, 2003; see Fig. 2A) using a microneedle. Images were acquired using a Nikon IX-70 inverted microscope fitted with a cooled CCD camera (Photometrics CH250), and a Ludl motorized XYZ stage and heating insert (Medical Systems). Electronic shutters regulated the fluorescence illumination. Image acquisition and processing used Inovision software. A field in each slice was selected that retained good morphology and had fluorescently labeled migrating cells adjacent to the DiI crystals. In a typical experiment, nearly all of the DiI-treated slices displayed appropriate labeling. The slices were illuminated with either a mercury or halogen lamp. A tetramethyl rhodamine isothiocyanate filter cube was used to observe the DiI-labeled cells. Fluorescence images were recorded every 5 min using a 10 × objective. Images were typically captured over 3–10 h using 0.015–0.20 s exposures. The time-lapse movies (Supplementary material, Videos S1–S3) were analysed for cells that migrated to the OB along the RMS. Tissue slices that showed migrating cells labeled with DiI were selected and cultured in CCM1 medium containing function-blocking peptide (CRLAQDECDVTEYC) that mimicked the disintegrin loop of ADAM2 (Bigler et al., 2000). For the negative controls, ADAM2 peptide with scrambled sequence (CETADYQRVECLDC) was used.
Fig. 2.
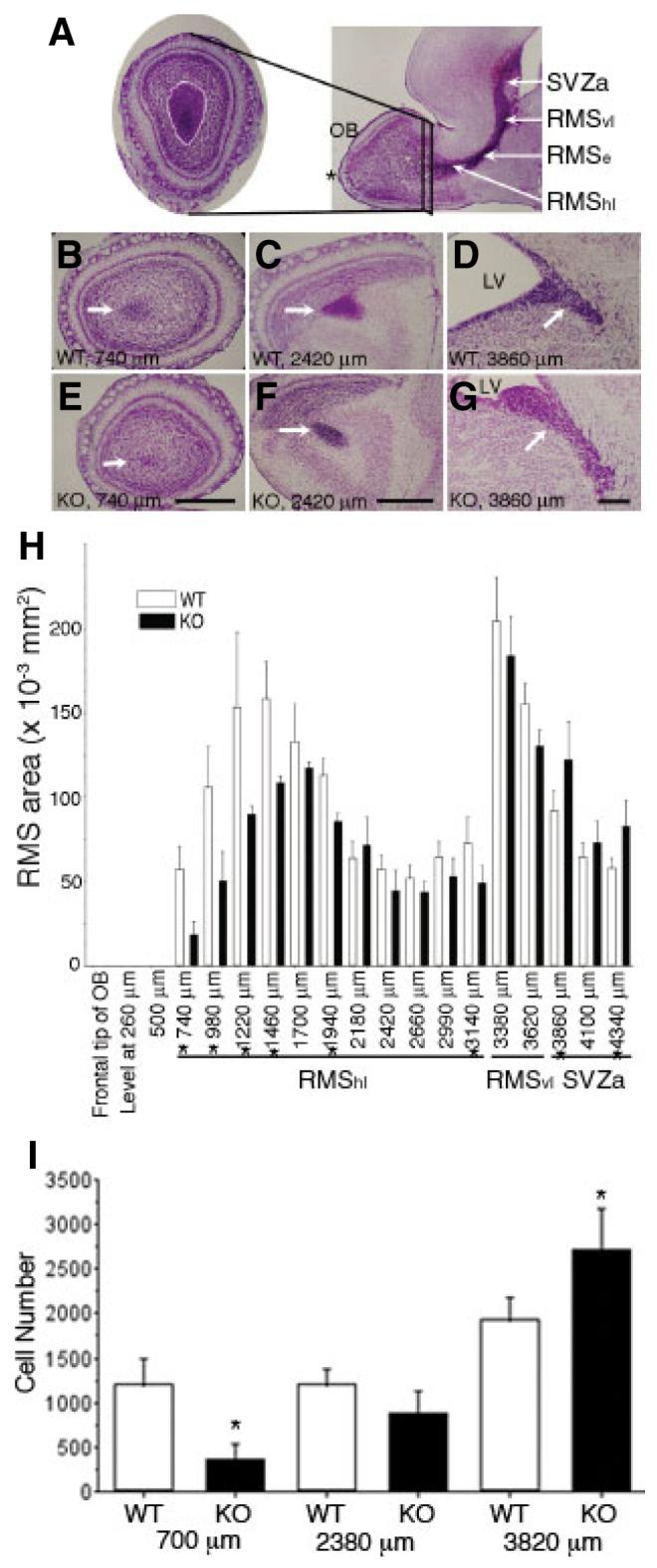
Altered histoarchitecture of the rostral migratory stream (RMS) of ADAM2 knockout (KO) mice (A) The RMS is distinguished by the intense Nissl staining in this parasagittal view of a P10 mouse forebrain (right). The asterisk indicates the frontal tip of the olfactory bulb (OB). The area of the RMS from wild-type (WT) and KO P10 mice was estimated from 20-μm-thick Nissl-stained, serial frontal sections derived from the frontal tip of the OB to the anterior part of the subventricular zone (SVZa; left). Areas of the RMS (demarcated by the white line) were measured on these frontal sections at 240 μm intervals. The RMS starts at the SVZa, then descends as the vertical limb (RMSvl), and finally becomes the horizontal limb (RMShl) near the OB. The elbow of the RMS (RMSe; Pencea & Luskin, 2003) is the junction between the RMSvl and the RMShl. (B—D) WT. Representative sections at the levels of 740 μm (end portion of the RMS), 2420 μm (RMShl, which denotes the horizontal limb region of the RMS near the OB) and 3860 μm (SVZa) from the frontal tip of the OB are shown. The RMS or SVZa (arrows) are characterized by a high cellular density at the center of the OB, or adjacent to the lateral ventricle (LV), respectively. (E—G) KO. Sections of the same level shown in (B—D). In each photograph (B—G), the right side is toward the dorsal part of the forebrain. Scale bar: 500 μm. (H) The RMS areas from the WT and the KO were compared between the same levels from the frontal tip of the OB to the SVZa. Each value represents the mean ± SD (N = 4 from four mice). The differences between the area of WT and KO at the levels (indicated by asterisks) were significant (P < 0.05). (I) The number of polysialic acid (PSA)-positive cells was counted in the RMS of WT and KO mice, and compared at the same distance from the tip of the OB to the SVZa. Each value represents the mean ± SD (N = 4 from four mice). The differences in cell number at the 700 μm and 3820 μm levels are significant (indicated by asterisks, P < 0.05).
Matrigel experiment for chain migration assay
After making sagittal sections of P5 mice forebrains, the RMSe (see Fig. 2A) was cut into square pieces of 100–200 μm in diameter and embedded in ice-cooled BD Matrigel™ Basement Membrane Matrix (growth factor reduced type, BD Biosciences, Lexington, KY, USA) diluted with CCM1 medium (Wichterle et al., 1997). The Matrigel containing pieces of RMS tissue were allowed to congeal in a culture dish and cultured in a humidified, 5% CO2, 37 °C incubator for 24 h. The migrating neuroblasts from the RMS explants were recorded over the next 3 h with differential interference contrast optics (20 × objective). For some experiments a function-blocking peptide that mimicked the disintegrin loop of ADAM2 was added to the culture medium at 19 h after explanting the RMS tissue, then the explants were incubated with the peptide for 5 h, after which migration was recorded over the next 3 h. For a negative control, a peptide with a scrambled sequence for the ADAM2 disintegrin loop was used.
Migration parameters
The time-lapse movies were analysed for net migration speed and direction. The center of soma of migrating cells were traced at 5-min intervals, and the speed and direction of migration during time-lapse recording were determined as described previously (Murase & Horwitz, 2002). Each value represents the mean and SD. Statistical analysis was performed by one-way analysis of variance with Scheffé’s multiple comparison procedure, or by unpaired t-test (significance of P < 0.05).
Bromodeoxyuridine (BrdU) immunohistochemistry
BrdU (Sigma; 10 mg/mL in sterile saline with 0.007 N NaOH) was administered intraperitoneally at a concentration of 50 mg/kg BW. For evaluation of cell proliferation, BrdU was injected 2 h before killing. For the extent of cell migration, BrdU was injected 48 h before killing.
Morphometric analysis
A contour of the RMS was determined by its high cellular density, and the RMS area was calculated from each section using Adobe Photoshop. The density of BrdU-positive cells was evaluated by comparing the number of BrdU-positive cells to the surfaces occupied by these cells with the aid of Nissl staining of adjacent sections. Apoptotic cells were visualized using anti-single-stranded DNA antibody (DakoCytomation, A450; Frankfurt et al., 1996), and the number of the apoptotic cells was counted on each frontal section. To count the number of TH- or calbindin-positive cells, frontal sections of the OB were immunostained by each antibody and the number of immunoreactive profiles counted in ADAM2 KO and WT sections.
Results
ADAM2 is expressed in the RMS
We first investigated the expression of ADAM2 in mouse brain. Figure 1A and B shows Nissl-stained sagittal sections containing the entire RMS and ADAM2 protein localization, respectively, in a P5 mouse. ADAM2 expression in the RMS was determined using a monoclonal antibody (Fig. 1B) and two independent polyclonal antibodies (not shown). Figure 1C shows the expression of ADAM2 mRNA in the RMS by in situ hybridization. The mRNA for ADAM2 was expressed throughout the RMS and was also present in the GCL but at a reduced level. A sense probe control for ADAM2 gave no signal (Fig. 1D). In the ADAM2 KO, neither antisense nor sense probes showed significant signals (Fig. 1E and F). These in situ hybridizations were confirmed by immunofluorescence using antibodies specific for ADAM2 or PSA, a marker for the migrating neuroblasts in the RMS (Rousselot et al., 1995). Co-localization of the staining patterns showed that the migrating cells express ADAM2 (Fig. 1G and H). Weak immunoreactivity was also seen in the GCL of the OB, suggesting that some granule cells and/or radially migrating neuroblasts retain ADAM2 protein. ADAM2 protein was not observed in the RMS of ADAM2 KO mice (Fig. 1I), however, PSA was expressed (Fig. 1J). Higher magnification of a tissue section stained with anti-PSA and anti-ADAM2 antibodies showed that ADAM2 protein was expressed on the membrane of RMS neuroblasts (Fig. 1K and L). Tissue sections dually stained with anti-GFAP or anti-tenascin-C and anti-ADAM2 antibodies showed that glial elements did not express ADAM2 (Fig. 1M, N, P and Q). In addition, anti-neurofilament antibody did not react with ADAM2-expressing neuroblasts (Fig. 1O and R).
Fig. 1.
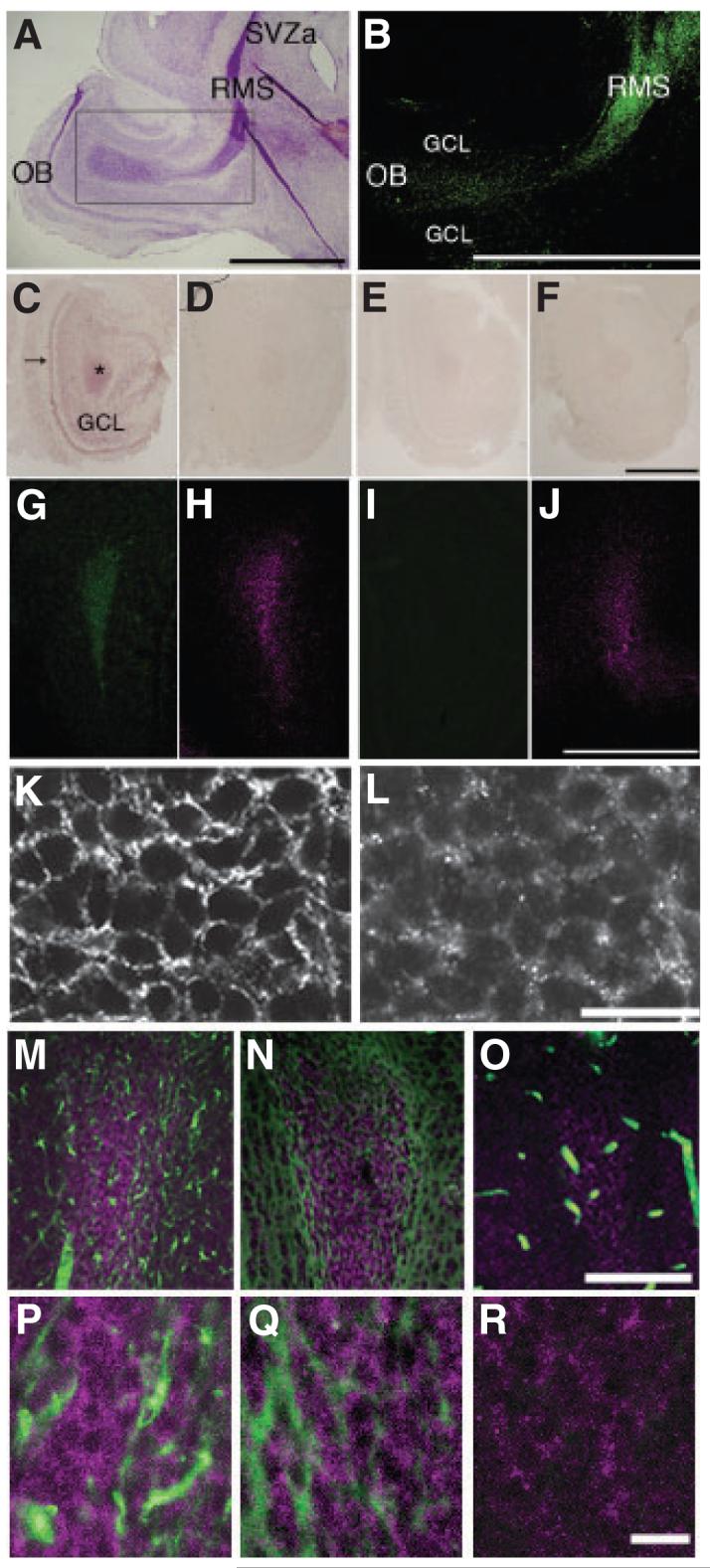
ADAM2 expression in the rostral migratory stream. (A) Nissl staining of a sagittal section from a postnatal day 5 (P5) mouse forebrain. The rostral migratory stream (RMS), which is characterized by high cellular density and intense Nissl staining, begins at the anterior portion of the subventricular zone (SVZa) and ends at the center of the olfactory bulb (OB). (B) ADAM2 immunoreactivity in the RMS. This is the adjacent section to that shown in (A, rectangular area), stained with 9D2.2, an anti-ADAM2 antibody. (C) ADAM2 mRNA expression in a frontal section (horizontal limb region; see legend for Fig. 2) from a P10 wild-type (WT) OB. (D) An adjacent section to that in (C) incubated with ADAM2 sense probe. (E) A frontal section of the OB from P10 ADAM2 knockout (KO) mouse incubated with ADAM2 antisense probe. (F) A section adjacent to (E) was incubated with the sense probe. (G and H) Frontal sections (vertical limb region; see Fig. 2 legend) of the OB from P10 WT mice were incubated with 9D2.2, anti-ADAM2 (green) and anti-polysialic acid (PSA) antibodies (magenta), respectively. (I and J) Frontal sections of the OB from P10 ADAM2 KO were incubated with anti-ADAM2 (green) and anti-PSA antibodies (magenta). GCL, granule cell layer. Scale bars: 500 μm. (K and L) Increased magnification of frontal sections from the WT RMS co-stained with anti-PSA and anti-ADAM2 (AB19030) antibodies. Scale bar: 10 μm. (M) Frontal section of the RMS from P21 WT mice co-stained with anti-glial fibrillary acidic protein (GFAP; green) and anti-ADAM2 antibodies (AB19030, magenta). (N) Frontal section of the RMS from P21 WT mice co-stained with anti-tenascin-C (green) and anti-ADAM2 antibodies (AB19030, magenta). (O) Frontal section of the RMS from P21 WT mice co-stained with anti-neurofilament monoclonal antibody (green) and anti-ADAM2 antibody (AB19030, magenta). Blood vessels containing IgG showed nonspecific staining for the secondary anti-mouse IgG antibody, but no specific staining for neurofilaments. Scale bar: 100 μm. (P) High magnification of (M). (Q) High magnification of (N). (R) High magnification of (O). Scale bar: 10 μm.
The RMS and SVZa from ADAM2 KO mice show altered morphologies
ADAM2 KO mice generated by conventional gene targeting methods survive to adulthood (Cho et al., 1998). We did not detect significant differences in body weight, brain weight or the ratio of brain weight to body weight among a sample of > five different P30, WT and KO mice (data not shown). In addition, the average weights of WTand KO mice at P0 were 1.56 g (N = 6) and 1.62 g (N = 6), respectively; a statistically insignificant difference. The size of the OB from serial frontal sections of the forebrains from P30 WT and KO mice also revealed no significant differences (data not shown). Next, we measured the area of the RMS from Nissl-stained frontal serial sections from P10 mice. Figure 2A shows a representative frontal section of the OB where the RMS is centrally located, and one sagittal section of the forebrain showing an entire contour of the RMS. The sagittal section also shows the name of each compartment of the RMS, e.g. the vertical limb (RMSvl), RMSe and horizontal limb (RMShl; Pencea & Luskin, 2003). Overall, the rostral portion of the RMS was thinner in the KO than in the WT mice. For example, the RMS observed at 740 μm from the frontal tip of the OB was significantly smaller (Fig. 2E) in the KO than in WT mice (Fig. 2B). In addition, the Nissl-stained region of the SVZa in the KO occupied a larger area (Fig. 2G) than in the WT (Fig. 2D), suggesting that increased numbers of neuroblasts remain in this region. Finally, we compared the area of the RMS and the PSA-positive cell number from WT and KO mice (Fig. 2H and I). All of the above observations were made on four individual WT and KO mice. They suggest that the RMS from ADAM2 KO mice has impaired neuroblast migration resulting in hypodevelopment of the rostral RMS and enlargement of the SVZa at P10.
ADAM2 affects the size of the OB and numbers of TH- or calbindin-positive interneurons
The hypodevelopment of the rostral part of the ADAM2 KO RMS might result in an altered size of the OB. To test this, we calculated the volume of the OB from serial frontal sections of forebrains from P10 mice, the stage from which we showed altered morphologies of the RMS in the KO in Fig. 2. No significant difference of the OB volume between WT and KO was observed at P10 (data not shown). In P0 WT, a frontal section 460 μm from the frontal tip of the OB showed robust RMS—GCL (Fig. 3A). As the RMS is not clearly distinguished from adjacent GCL in this stage, we use the term RMS—GCL to describe the area containing the migrating neuroblasts at P0. As shown in Fig. 2, describing the P10 RMS area, the RMS—GCL area (460 μm level) from P0 KO (Fig. 3D) was smaller than that from P0 WT (Fig. 3A). However, at the 1520 μm level, WT RMS—GCL (Fig. 3C) appeared slightly smaller than KO RMS—GCL (Fig. 3F), suggesting retention of the neuroblasts in the caudal part of the RMS to SVZa. Furthermore, the volume of the KO OB decreased to 76% of the WT OB (Fig. 3G). These observations suggest that deletion of the ADAM2 gene affects the size of the OB by inducing hypodevelopment of the rostral part of the RMS at early developmental stages (e.g. P0) but not at later postnatal stages.
Fig. 3.
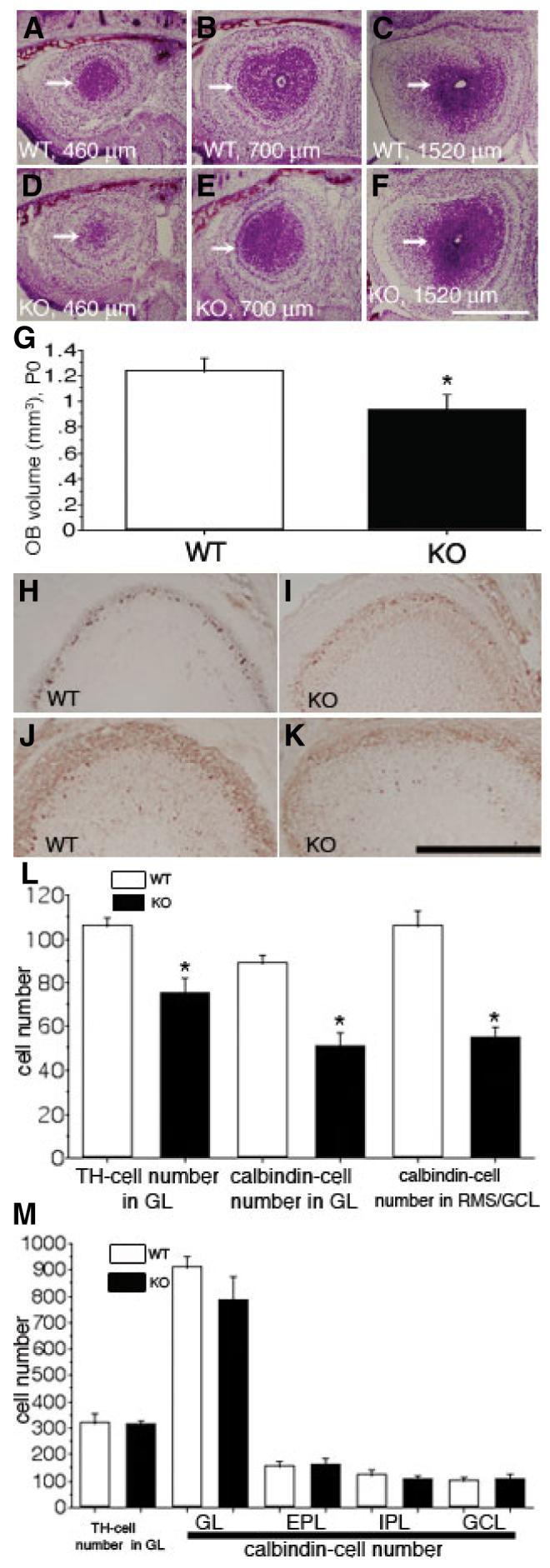
ADAM2 affects the size of the olfactory bulb (OB) at an early developmental stage. (A—C) Wild-type (WT) mice at P0. Representative, Nissl-stained sections at the level of 460 μm, 700 μm and 1520 μm from the frontal tip of the OB are shown; rostral migratory stream (RMS)—granular cell layer (GCL) (arrows). (D—F) Knockout (KO) mice at P0. Sections are at the same levels shown in (A—C). (G) The OB volume from the WT and the KO were compared. Each value represents the mean ± SD (N = 5 from five mice); the difference in volume of the OB between WT and KO is=significant (indicated by asterisk, P < 0.05). (H and I) WT (H) and KO (I) OB from P0 mice stained with an anti-tyrosine hydroxylase (TH) antibody. (J and K) WT (J) and KO (K) OB from P0 mice stained with an anti-calbindin antibody. (L) The difference in the numbers of TH- and calbindin-positive cells from the WT and KO mice at P0 are significant (indicated by asterisks, P < 0.05); each value represents the mean ± SD (N = 4 from four mice). (M) The numbers of TH-and calbindin-positive cells from the WT and KO mice at P30 were compared. Each value represents the mean ± SD (N = 4 from four mice), and is not statistically significantly different. EPL, external plexiform layer; GL, glomerular layer; IPL, internal plexiform layer. Scale bar: 500 μm (A—F); 250 μm (H—K).
We also measured the number of TH-positive periglomerular cells and of calbindin-positive periglomerular cell subset and interneurons in the GCL to determine whether there is a decrease in the number of interneurons of the OB in ADAM2 KO mice. The number of TH- or calbindin-positive cells was lower in ADAM2 KO OB at P0 (Fig. 3H–K). TH-positive cells were found only in the glomerular layer (Fig. 3H and I), whereas calbindin-positive cells were observed both in the glomerular layer and RMS—GCL (Fig. 3J and K). At 840 μm from the frontal tip of the OB, the number of immunoreactive cells was counted and compared between the WT and KO (Fig. 3L). The numbers of TH- or calbindin-positive cells were lower both in the glomerular layer and in the RMS—GCL at P0. However, the numbers of TH- or calbindin-positive cells from the sections of OB (at the level of 1600 μm) from P30 mice were not statistically different between WT and KO (Fig. 3M).
Neuroblasts in the RMS show altered migration
Hypodevelopment in the rostral region of the RMS at P0 (Fig. 3) and P10 (Fig. 2) suggests either a migratory insufficiency, decreased proliferation or accelerated cell death of neuroblasts in ADAM2 KO mice. To address these possibilities, we compared the number of proliferating and dying cells in the RMS from P10 WT and KO mice using BrdU labeling in vivo. BrdU was injected into the peritoneal space and, after 2 h, the mice were killed and the brains sectioned frontally along the RMS. Immunostaining for BrdU showed the number and location of proliferating cells in the RMS, and immunohistochemistry, with an anti-single-stranded DNA antibody, detected the number and location of dying cells. These two parameters did not differ detectably in the rostral region of the RMS from WT and KO mice, indicating that the ratio of proliferating and dying cells in the RMS was normal in KO mice (Fig. 4A and B).
Fig. 4.
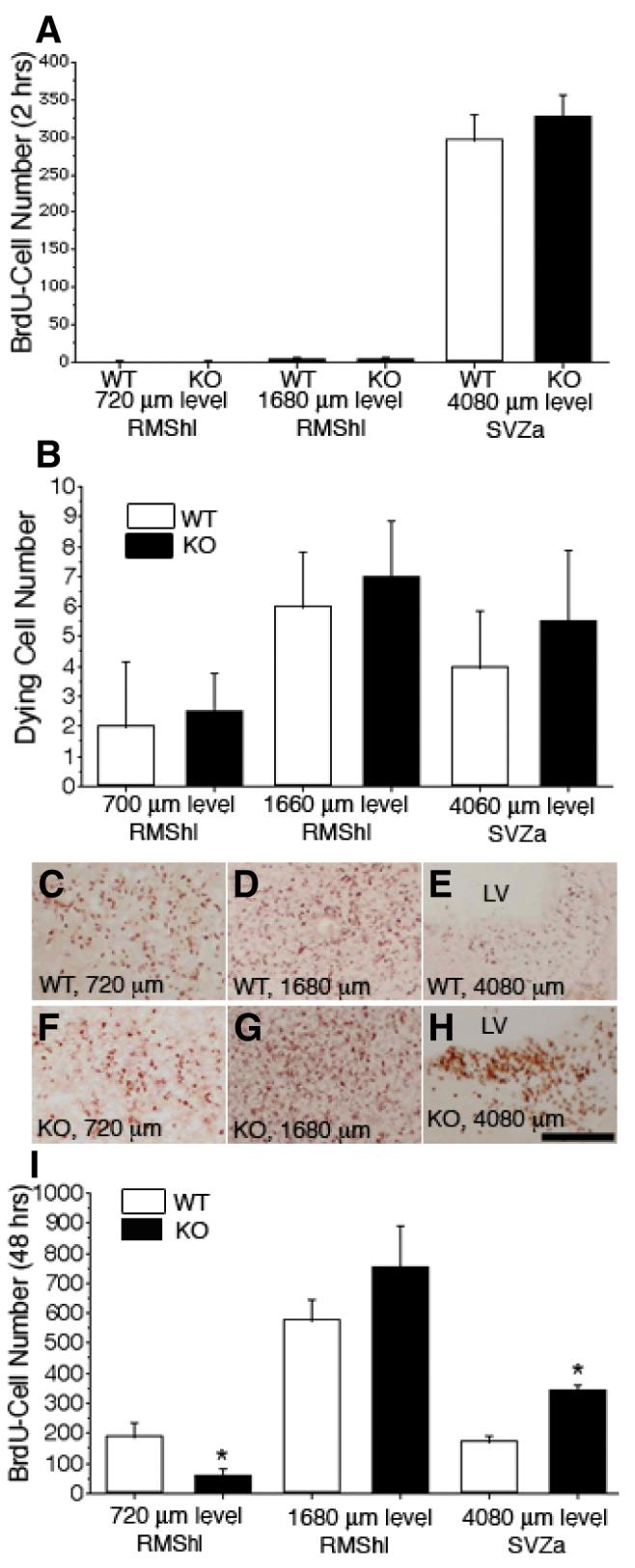
Delayed tangential migration in the ADAM2 knockout (KO) rostral migratory stream (RMS). P10 postnates were injected with bromodeoxyuridine (BrdU), killed 2 h later, and then fixed for BrdU and stained with an anti-single-stranded DNA antibody to evaluate proliferative activity and cell death of neuroblasts, respectively. Neuroblasts migration in vivo was estimated in P8 postnatal mice by injecting BrdU, killing 48 h later and fixing for BrdU immunohistochemistry. In both wild-type (WT) and KO P10 mice, serial frontal sections (20 μm thickness each) from the frontal tip of the olfactory bulb (OB) to the anterior part of the subventricular zone (SVZa) were collected and stained with an anti-BrdU antibody or anti-single-stranded DNA antibody. The number of BrdU-positive cells (BrdU-cells) was evaluated on sections at 240 μm intervals. In each image, the right side is toward the dorsal part of the forebrain. (A) Neuroblast proliferative activity was estimated from the number of BrdU-cells (2 h after injection) from WT and KO mice at the same distance between the frontal tip of the OB and the SVZa. Each value represents the mean ± SD (N = 4 from four mice); there are no statistical differences (P < 0.05). (B) The number of dying neuroblasts in the RMS from WT and KO mice was compared at the same distance between the frontal tip of the OB and the SVZa. Each value represents the mean ± SD (N = 4 from four mice); there were no statistically significant differences (P <= 0.05). (C—E) WT. Representative sections at the end portion of the RMS (720 μm level), horizontal limb of the RMS (RMShl; 1680 μm level) and SVZa (4080 μm level) from the frontal tip of the OB are shown. BrdU-cells (48 h after injection) were concentrated in the RMS. At the SVZa level in (E), few BrdU-cells are seen. (F—H) KO. Sections are at the same levels shown in (C—E). In (F), the BrdU-cell density (48 h after injection) is low compared with WT (C). At the 1680 μm level, the section from the KO mice shows significant numbers of BrdU-cells (G), and at the SVZa level, many BrdU-cells are seen (H). (I) The number of BrdU-cells (48 h after injection) from WT and KO mice were compared at the same levels from the frontal tip of the OB to the SVZa. At the rostral part of the RMS, the KO mice show fewer BrdU-cells than that from WT mice; however, at the mid portion between the RMShl and the SVZa, KO mice have more cells than WT. Each value represents the mean ± SD (N = 4 from four mice); the differences at the 720 μm and 4080 μm levels are significant (indicated by asterisks, P < 0.05). LV, lateral ventricle; RMShl, horizontal limb region of the RMS. Scale bar: 500 μm.
We next evaluated neuroblast migration by determining the number and location of BrdU-positive cells (BrdU-cells) in frontal serial sections of the RMS, from the tip of the OB to the SVZa, 48 h after BrdU injection. The 48 h latency was used to allow migration of neuroblasts that are generated and labeled primarily in the SVZa (Saghatelyan et al., 2004). We observed many BrdU-positive cells at the 720 μm and 1680 μm levels in the RMS from P10 WT mice; this number was decreased slightly at the 4080 μm level in the SVZa (Fig. 4C–E). In contrast, the number of BrdU-cells in the KO RMS was 30% that of the WT at the 720 μm level. However, at 1680 μm and 4080 μm levels, the number of BrdU-cells in the KO was higher (144% and 183%, respectively) than that of the WT (Fig. 4F–H). The decreased number of BrdU-cells in the rostral part of the RMS is consistent with the Nissl staining in Fig. 2 and suggests decreased neuroblast migration in the ADAM2 KO. The increased number of BrdU-cells in the caudal part of the RMShl, the RMSvl and the SVZa suggests retention of neuroblasts in these regions and is consistent with decreased migration for the KO neuroblasts.
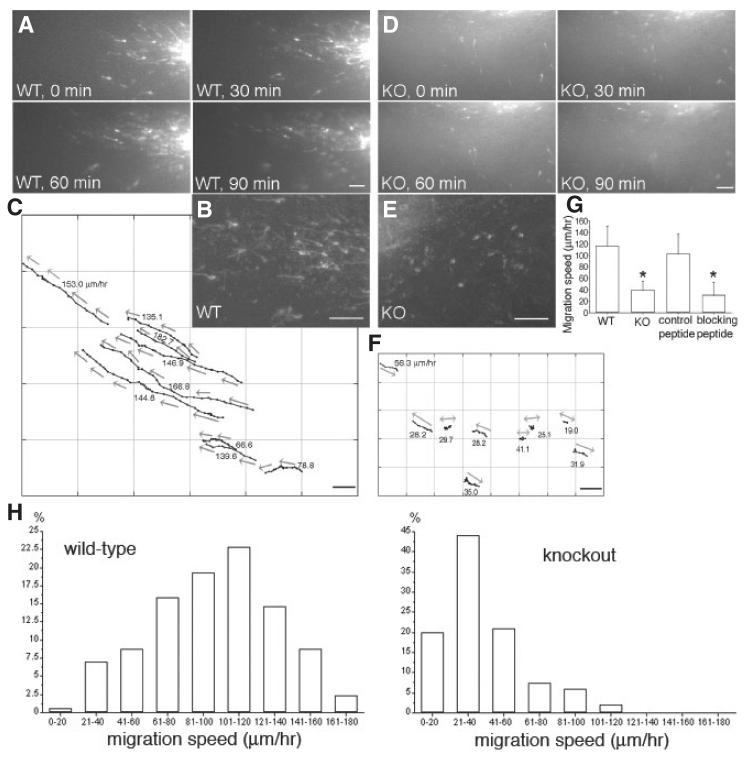
Finally, we assayed migration rates directly using 200-μm living slices containing the RMS. We have used this preparation previously to visualize and quantify neuroblast migration at high spatial and temporal resolution (Murase & Horwitz, 2002). In brief, slices are made from sagittal sections of forebrains from P10 mice. A crystal of DiI is added to the RMSe (see Fig. 2A) and, after a 5-h incubation, images of migrating cells are captured for 3–4 subsequent hours. As shown in Fig. 5A–C and supplementary Video S1, the overall direction of RMS migration in slices from WT was highly persistent and directed toward the OB. The migrating cells were highly polarized with long leading processes and relatively small somata. This morphology is similar to that observed by silver impregnation (Kishi, 1987). The average speed of migration in WT slices was 116 μm/h, which is similar to the values reported in our previous study (Murase & Horwitz, 2002). Using this in situ system, we assayed migration in slices from ADAM2 KO mice (Fig. 5D–F). The migrating neuroblasts in the KO RMS (Fig. 5E) were morphologically distinguishable from those in the WT RMS (Fig. 5B), displaying shorter leading processes (WT, 27 ± 3 μm; KO, 17 ± 2 μm, statistically significant by unpaired t-test, significance of P < 0.05). In addition the net migration rates were slower. Migration trajectories of the KO neuroblasts showed a less directed pathway as they often migrated bidirectionally, which resulted in shorter net migratory distances (neuroblasts marked by double-headed arrows in Fig. 5F). The migration speed of cells in the ADAM2 KO decreased to ∼40% of the control values (P < 0.05; Fig. 5G).
Fig. 5.
Disturbed tangential migration of ADAM2 knockout (KO) neuroblasts. Brain slices from a P10 mouse [wild-type (WT) or KO] were labeled with DiI and their migration recorded for 3–4 h. (A) Time-lapse sequence of WT neuroblasts migrating from the anterior part of the subventricular zone (SVZa; right bottom corner) toward the olfactory bulb (OB; left upper corner). The interval between each image is 30 min. See supplementary Video S1. (B) Higher magnification image of DiI-labeled WT neuroblasts migrating toward the OB. (C) Graphical representation of the migration of the neuroblasts in (A). Each point represents the position of the cell body at 5-min intervals. Arrows show the direction of neuroblast migration. (D) Time-lapse sequence of ADAM2 KO neuroblasts migrating from the SVZa (right) toward the OB (left). The interval between each image is 30 min. See supplementary Video S2. (E) Higher magnification image of DiI-labeled ADAM2 KO neuroblasts. (F) Graphical representation of the migration of the ADAM2 KO neuroblasts in (D). Each point represents the position of the cell body at 5-min intervals. Neuroblasts without a clear direction were marked with double-headed arrows. (G) Reduced migration rate of RMS neuroblasts from ADAM2 KO mice or in the presence of a function-blocking ADAM2 peptide. Each value is the mean ± SD. WT (N = 46, five slices from five mice), ADAM2 KO (N = 36, five slices from five mice), control peptide (N = 31, four slices from four mice), blocking peptide (N = 30, five slices from five mice). Statistically significant data (P < 0.05) are indicated by asterisks. Scale bars: 50 μm. (H) Distribution of migration speeds of the RMS neuroblasts from WT (N = 171, six slices from six mice) and KO (N = 202, six slices from six mice)mice.
We tested the acute effect of a peptide that contains the disintegrin loop on neuroblast migration in WT slices. The peptide inhibited the net migration speed to ∼30% of control (Fig. 5G). In addition to the decrease in migration speed, the direction of movement was also perturbed by the peptide, and many cells did not move unidirectionally toward the OB (see supplementary Video S3). We then analysed the distribution of neuroblast migration speeds from both the WT and KO RMS (Fig. 5H). The mode speed (most common) for each phenotype is ∼101–120 μm/h (WT) and ∼21–40 μm/h (KO), and the mean speed for each phenotype is 116 μm/h (WT) and 38 μm/h (KO), respectively. In the KO, we observed a small number of rapidly migrating neuroblasts (> 60 μm/h). Also, in the WT 20% of neuroblasts migrated slowly.
Migration of RMS cells from ADAM2 KO is perturbed in culture
In the above experiments, we showed that ADAM2 promotes directed migration in the RMS. We next probed a mechanism by which ADAM2 may contribute to neuroblast migration. The culture of neuroblasts in Matrigel promotes cell—cell interactions, which result in a chain-like appearance (Wichterle et al., 1997). Neuroblasts from WT mice migrated away from the original RMS explant and formed cell aggregations resembling chains (Fig. 6A; supplementary Video S4). Each neuroblast has a long leading process, as they do in situ; however, migration in this system is not as homogeneous and unidirectional as in vivo. Many neuroblasts migrate out of the explant, but migration is not as unidirectional as in the slice cultures. Isolated cells were rarely seen, instead nearly all of the neuroblasts were in contact with other neuroblasts.
Fig. 6.
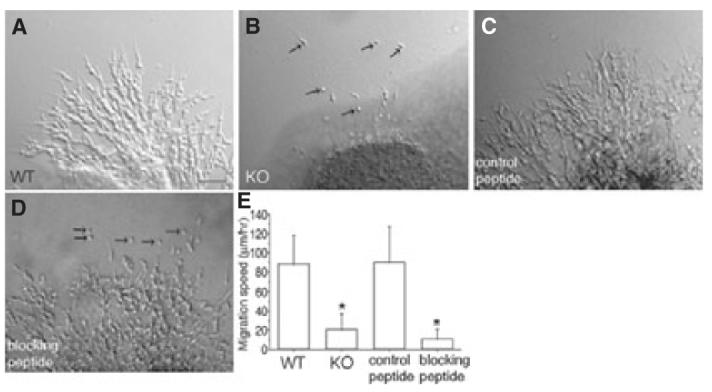
Perturbed chain migration of neuroblasts in explant cultures from ADAM2 knockout (KO) or wild-type (WT) mice treated with an ADAM2 function-blocking peptide. Rostral migratory stream (RMS) tissue from ADAM2 KO or WT mice was cultured in Matrigel. The tissue from WT mice was cultured either in the absence or presence of adhesion-blocking peptides. The migration was analysed from differential interference contrast images. For the peptide-blocking experiments (C and D), the explants were cultured for 24 h before addition of the peptides. (A) Neuroblasts migrating away from an RMS explant from WT mice embedded in Matrigel. See supplementary Video S4. Scale bar: 50 μm. (B) Cells from the RMS of KO mice migrate as single cells (arrows). See supplementary Video S5. (C) A peptide whose sequence contains the scrambled amino acids in the ADAM2 disintegrin loop was added to Matrigel cultures of the RMS from WT mice, as a control peptide. See supplementary Video S6. (D) Function-blocking peptide whose sequence corresponds to the ADAM2 disintegrin loop was added into WT Matrigel culture. Isolated neuroblasts (arrows) are seen. See supplementary Video S7. (E) Reduced net migration rate of RMS neuroblasts either from ADAM2 KO, or from WT mice and incubated with a function-blocking ADAM2 peptide. WT (N = 34, three explants from three mice), ADAM2 KO (N = 42, three explants from three mice), control peptide (N = 37, three explants from three mice), blocking peptide (N = 37, three explants from three mice). The differences in migration speed between WT and KO, and control peptide and blocking peptide are significant (P < 0.05; indicated by asterisks).
In contrast, fewer cells migrated out of the ADAM2 KO explant, and those that did migrated more slowly and tended to be present as single neuroblasts that were not in contact with other neuroblasts (Fig. 6B; supplementary Video S5). The migration speeds decreased to 23% of the control values (P < 0.05; Fig. 6E). When a blocking peptide was added to the WT explant cultures (in which neuroblasts had been allowed to migrate out of the explants for 24 h before addition of the peptide), the migration speed of neuroblasts decreased to 12% of control slices incubated with a scrambled sequence peptide (P < 0.05; Fig. 6C–E; supplementary Video S6). In addition, the groups of cells appeared less organized and some neuroblasts appeared as single cells (Fig. 6D; supplementary Video S7). Together, these data suggest a role for ADAM2 in chain migration in explant cultures.
Discussion
This study was motivated by the presence of ADAM2 mRNA in the brain (Karkkainen et al., 2000). We have extended this observation by demonstrating the presence of ADAM2 in the RMS, the defined pathway along which neuroblasts migrate from the SVZa to the OB, and by showing that it contributes to normal neuroblast migration along this pathway. We have shown that ADAM2 protein is exclusively expressed by neuroblasts in the RMS. Analysis of ADAM2 KO mice shows inhibited migration and hypodevelopment of the rostral RMS. In addition, the OBs of ADAM2 KO mice are significantly smaller than those from WT mice at birth. These morphological changes in vivo are in accord with the perturbed directional migration of neuroblasts observed in slices from KO and in WT mice incubated with a function-blocking peptide. Analyses of RMS cells migrating out of explants in Matrigel suggest that the inhibition may arise, at least in part, from altered neuroblast cell—cell interactions.
Neuroblasts in the RMS undergo tangential migrations that are not guided by glia (Kishi et al., 1990). In contrast to the glial-guided radial migrations in the CNS (Rakic, 2006), relatively less is know about the mechanisms that guide tangential cellular migrations including those in the RMS. The PSA moiety on neural cell adhesion molecule (NCAM) has been implicated in RMS migration, presumably by reducing cell—cell adhesion (Fujimoto et al., 2001). In PSA—NCAM KO mice (Tomasiewicz et al., 1993; Cremer et al., 1994; Ono et al., 1994; Chazal et al., 2000; Weinhold et al., 2005), for example, the altered interactions between neuroblasts and/or surrounding astrocytes, produced by the removal of the large, charged carbohydrates, inhibits normal migration of the neuroblasts. This results in their accumulation in the RMS and a consequent decrease in the size of the adult OB. Thus, PSA appears to function as a negative regulator of cell—cell interactions. However, PSA-positive chains and their surrounding glial tubes are not morphologically apparent before P14, and begin to form after P15 (Tramontin et al., 2003; Peretto et al., 2005; Bonfanti, 2006). We and others have previously shown (Murase & Horwitz, 2002; 2004; Peretto et al., 2005) that PSA-immunoreactivity in the RMS is weak before P14. Considering the paucity of PSA-immunoreactivity before P14, other cell surface molecules including ADAM2 likely mediate rapid RMS migration. The fact that we did not observe significant morphological differences in the RMS and the OB cytoarchitecture between WT and ADAM2 KO in the adult mice suggests that ADAM2 function is important primarily at early developmental stages. The isolated disintegrin domain of the ADAM2 can interact with integrins (Tomczuk et al., 2003), but there is currently no evidence for a physiological function for this interaction (He et al., 2003). Nonetheless, because recent studies reported that β1 integrin subunit regulates the migration speed and chain formation of the RMS neuroblasts (Murase & Horwitz, 2002; Belvindrah et al., 2007), the possibility that ADAM2 is mediating its effect on cell migration through modulating integrin activity would remain as a next important question.
Recently, retention of neuroblasts in the SVZa and delayed RMS migration were also reported in doublecortin KO mice. The morphology of the RMS was altered, although not as dramatically as in the NCAM KO; in contrast to NCAM KO mice, no significant differences were reported for the size of the OB (Koizumi et al., 2006).
The phenotype that we have observed for ADAM2 KO mice shares features with both of these mice. In particular, a significant inhibition of migration was seen in slice cultures and Matrigel assays. Interestingly, in slice cultures both the doublecortin and ADAM2 KO mice showed alterations in the leading processes, although the details differed. Loss of doublecortin resulted in highly branched leading processes, while the processes were shorter on neuroblasts from the ADAM KO mice.
In vivo, the development of the OB was perturbed in ADAM2 KO mice at P0, when the peak of neurogenesis for the OB is beginning (Bayer, 1983). However, this phenotype was not apparent at later stages (P10) or in the adult, pointing to the contributions of other molecules and/or adaptation at these later stages. The distribution of migration speeds of neuroblasts from WTand KO mice reveals that even in the KO RMS ∼20% of the neuroblasts migrated with velocities > 60 μm/h. Thus, while the average speed of the two phenotypes is very different, a significant number of cells migrate rapidly even in the KOs.
In the doublecortin KO a statistically significant reduction in the size of the OB was not seen. In contrast, NCAM KO mice showed a decreased size of the OB at birth that persisted in the adult (Tomasiewicz et al., 1993). Hence, both the in vitro and in vivo phenotypes of ADAM2 KO mice are more similar to those of doublecortin rather than NCAM KO mice.
All of the data therefore are consistent with ADAM2 regulating migration in the RMS. The mechanism by which it does so is unclear. One possibility is through altered cell—cell interactions. Because the protease domain of ADAM2 is inactive (Wolfsberg et al., 1993), adhesion influenced by the disintegrin loop may be involved. In addition, ADAM2 may regulate, either directly or indirectly, signals that support a polarized morphology and migration, as inhibition of ADAM2 leads to cells with shorter processes.
Thus, ADAM2 appears to facilitate migration along the RMS. Migration of luteinizing hormone-releasing hormone neurons from the olfactory placode to the forebrain (Schwanzel-Fukuda & Pfaff, 1989), precerebellar neurons to the brainstem (Yachnis & Rorke, 1999), and cerebral/thalamic γ-aminobutyric acid (GABA)ergic interneurons (Letinic & Rakic, 2001; Letinic et al., 2002), all are reported to employ neurophilic contacts as proposed for RMS neuroblasts. Migratory deficits of these neurons result in Kallman syndrome, Joubert syndrome and schizophrenia, respectively. However, the molecular mechanisms behind these migrations are largely unknown. ADAM2 participates in neurophilic migration of RMS neuroblasts, at least in vitro, and might therefore be involved in other neurophilic migrations as well.
Supplementary Material
Acknowledgements
The authors wish to thank D. G. Myles and P. Primakoff for ADAM2 knockout mice. This work was supported by National Institutes of Health Grants, GM23244 and U54 G64346 (Cell Migration Consortium) from NIGMS to A.F.H., and NIH GM48739 and March of Dimes grant FY05-140 to J.M.W. Initiation of this study was supported from Human Frontier Science Program Organization award to S.M.
Abbreviations
- ADAM
a disintegrin and metalloprotease
- BrdU
bromodeoxyuridine
- CCM
cell culture media
- DiI
1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate
- GCL
granular cell layer
- GFAP
glial fibrillary acidic protein
- KO
knockout
- NCAM
neural cell adhesion molecule
- OB
olfactory bulb
- PBS
phosphate-buffered saline
- PCR
polymerase chain reaction
- PSA
polysialic acid
- RMS
rostral migratory stream
- RMSe
elbow of the RMS
- RMShl
horizontal limb of the RMS
- RMSvl
vertical limb of the RMS
- SSC
standard saline citrate
- SVZa
anterior part of the subventricular zone
- TH
tyrosine hydroxylase
- WT
wild-type
Footnotes
Please note: Blackwell Publishing are not responsible for the content or functionality of any supplementary materials supplied by the authors. Any queries (other than missing material) should be directed to the correspondence author for the article.
References
- Altman J. Autoradiographic and histological studies of postnatal neurogenesis. IV. Cell proliferation and migration in the anterior forebrain, with special reference to persisting neurogenesis in the olfactory bulb. J. Comp. Neurol. 1969;137:433–457. doi: 10.1002/cne.901370404. [DOI] [PubMed] [Google Scholar]
- Aufderheide E, Ekblom P. Tenascin during gut development: appearance in the mesenchyme, shift in molecular forms, and dependence on epithelial—mesenchymal interactions. J. Cell Biol. 1988;107:2341–2349. doi: 10.1083/jcb.107.6.2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastianelli E, Pochet R. Calmodulin, calbindin-D28k, calretinin and neurocalcin in rat olfactory bulb during postnatal development. Brain Res. Dev. Brain Res. 1995;87:224–227. doi: 10.1016/0165-3806(95)00073-m. [DOI] [PubMed] [Google Scholar]
- Bayer SA. 3H-thymidine-radiographic studies of neurogenesis in the rat olfactory bulb. Exp. Brain Res. 1983;50:329–340. doi: 10.1007/BF00239197. [DOI] [PubMed] [Google Scholar]
- Belvindrah R, Hankel S, Walker J, Patton BL, Muller U. Beta1 integrins control the formation of cell chains in the adult rostral migratory stream. J. Neurosci. 2007;27:2704–2717. doi: 10.1523/JNEUROSCI.2991-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigler D, Takahashi Y, Chen MS, Almeida EA, Osbourne L, White JM. Sequence-specific interaction between the disintegrin domain of mouse ADAM 2 (fertilin beta) and murine eggs. Role of the alpha(6) integrin subunit. J. Biol. Chem. 2000;275:11576–11584. doi: 10.1074/jbc.275.16.11576. [DOI] [PubMed] [Google Scholar]
- Blobel CP, Wolfsberg TG, Turck CW, Myles DG, Primakoff P, White JM. A potential fusion peptide and an integrin ligand domain in a protein active in sperm-egg fusion. Nature. 1992;356:248–252. doi: 10.1038/356248a0. [DOI] [PubMed] [Google Scholar]
- Bonfanti L. PSA-NCAM in mammalian structural plasticity and neurogenesis. Prog. Neurobiol. 2006;80:129–164. doi: 10.1016/j.pneurobio.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Chazal G, Durbec P, Jankovski A, Rougon G, Cremer H. Consequences of neural cell adhesion molecule deficiency on cell migration in the rostral migratory stream of the mouse. J. Neurosci. 2000;20:1446–1457. doi: 10.1523/JNEUROSCI.20-04-01446.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho C, Bunch DO, Faure JE, Goulding EH, Eddy EM, Primakoff P, Myles DG. Fertilization defects in sperm from mice lacking fertilin beta. Science. 1998;281:1857–1859. doi: 10.1126/science.281.5384.1857. [DOI] [PubMed] [Google Scholar]
- Cremer H, Lange R, Christoph A, Plomann M, Vopper G, Roes J, Brown R, Baldwin S, Kraemer P, Scheff S, Barthels D, Rajewsky K, Wille W. Inactivation of the N-CAM gene in mice results in size reduction of the olfactory bulb and deficits in spatial learning. Nature. 1994;367:455–459. doi: 10.1038/367455a0. [DOI] [PubMed] [Google Scholar]
- Debus E, Weber K, Osborn M. Monoclonal antibodies specific for glial fibrillary acidic (GFA) protein and for each of the neurofilament triplet polypeptides. Differentiation; Res. Biol. Diversity. 1983;25:193–203. doi: 10.1111/j.1432-0436.1984.tb01355.x. [DOI] [PubMed] [Google Scholar]
- Dodd J, Morton SB, Karagogeos D, Yamamoto M, Jessell TM. Spatial regulation of axonal glycoprotein expression on subsets of embryonic spinal neurons. Neuron. 1988;1:105–116. doi: 10.1016/0896-6273(88)90194-8. [DOI] [PubMed] [Google Scholar]
- Doetsch F, Caille I, Lim DA, Garcia-Verdugo JM, Alvarez-Buylla A. Subventricular zone astrocytes are neural stem cells in the adult mammalian brain. Cell. 1999;97:703–716. doi: 10.1016/s0092-8674(00)80783-7. [DOI] [PubMed] [Google Scholar]
- Frankfurt OS, Robb JA, Sugarbaker EV, Villa L. Monoclonal antibody to single-stranded DNA is a specific and sensitive cellular marker of apoptosis. Exp. Cell Res. 1996;226:387–397. doi: 10.1006/excr.1996.0240. [DOI] [PubMed] [Google Scholar]
- Fujimoto I, Bruses JL, Rutishauser U. Regulation of cell adhesion by polysialic acid. Effects on cadherin, immunoglobulin cell adhesion molecule, and integrin function and independence from neural cell adhesion molecule binding or signaling activity. J. Biol. Chem. 2001;276:31745–31751. doi: 10.1074/jbc.M104525200. [DOI] [PubMed] [Google Scholar]
- Hack I, Bancila M, Loulier K, Carroll P, Cremer H. Reelin is a detachment signal in tangential chain-migration during postnatal neurogenesis. Nat. Neurosci. 2002;5:939–945. doi: 10.1038/nn923. [DOI] [PubMed] [Google Scholar]
- He ZY, Brakebusch C, Fassler R, Kreidberg JA, Primakoff P, Myles DG. None of the integrins known to be present on the mouse egg or to be ADAM receptors are essential for sperm-egg binding and fusion. Dev Biol. 2003;254:226–237. doi: 10.1016/s0012-1606(02)00043-x. [DOI] [PubMed] [Google Scholar]
- Huovila AP, Turner AJ, Pelto-Huikko M, Karkkainen I, Ortiz RM. Shedding light on ADAM metalloproteinases. Trends Biochem. Sci. 2005;30:413–422. doi: 10.1016/j.tibs.2005.05.006. [DOI] [PubMed] [Google Scholar]
- Karkkainen I, Rybnikova E, Pelto-Huikko M, Huovila AP. Metalloprotease-disintegrin (ADAM) genes are widely and differentially expressed in the adult CNS. Mol. Cell. Neurosci. 2000;15:547–560. doi: 10.1006/mcne.2000.0848. [DOI] [PubMed] [Google Scholar]
- Kishi K. Golgi studies on the development of granule cells of the rat olfactory bulb with reference to migration in the subependymal layer. J. Comp. Neurol. 1987;258:112–124. doi: 10.1002/cne.902580109. [DOI] [PubMed] [Google Scholar]
- Kishi K, Peng JY, Kakuta S, Murakami K, Kuroda M, Yokota S, Hayakawa S, Kuge T, Asayama T. Migration of bipolar subependymal cells, precursors of the granule cells of the rat olfactory bulb, with reference to the arrangement of the radial glial fibers. Arch. Histol. Cytol. 1990;53:219–226. doi: 10.1679/aohc.53.219. [DOI] [PubMed] [Google Scholar]
- Koizumi H, Higginbotham H, Poon T, Tanaka T, Brinkman BC, Gleeson JG. Doublecortin maintains bipolar shape and nuclear translocation during migration in the adult forebrain. Nat. Neurosci. 2006;9:779–786. doi: 10.1038/nn1704. [DOI] [PubMed] [Google Scholar]
- Leighton PA, Mitchell KJ, Goodrich LV, Lu X, Pinson K, Scherz P, Skarnes WC, Tessier-Lavigne M. Defining brain wiring patterns and mechanisms through gene trapping in mice. Nature. 2001;410:174–179. doi: 10.1038/35065539. [DOI] [PubMed] [Google Scholar]
- Letinic K, Rakic P. Telencephalic origin of human thalamic GABAergic neurons. Nat. Neurosci. 2001;4:931–936. doi: 10.1038/nn0901-931. [DOI] [PubMed] [Google Scholar]
- Letinic K, Zoncu R, Rakic P. Origin of GABAergic neurons in the human neocortex. Nature. 2002;417:645–649. doi: 10.1038/nature00779. [DOI] [PubMed] [Google Scholar]
- Lois C, Garcia-Verdugo JM, Alvarez-Buylla A. Chain migration of neuronal precursors. Science. 1996;271:978–981. doi: 10.1126/science.271.5251.978. [DOI] [PubMed] [Google Scholar]
- Luskin MB. Restricted proliferation and migration of postnatally generated neurons derived from the forebrain subventricular zone. Neuron. 1993;11:173–189. doi: 10.1016/0896-6273(93)90281-u. [DOI] [PubMed] [Google Scholar]
- Murase S, Horwitz AF. Deleted in colorectal carcinoma and differentially expressed integrins mediate the directional migration of neural precursors in the rostral migratory stream. J. Neurosci. 2002;22:3568–3579. doi: 10.1523/JNEUROSCI.22-09-03568.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myles DG, Kimmel LH, Blobel CP, White JM, Primakoff P. Identification of a binding site in the disintegrin domain of fertilin required for sperm-egg fusion. Proc. Natl Acad. Sci. USA. 1994;91:4195–4198. doi: 10.1073/pnas.91.10.4195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng KL, Li JD, Cheng MY, Leslie FM, Lee AG, Zhou QY. Dependence of olfactory bulb neurogenesis on prokineticin 2 signaling. Science. 2005;308:1923–1927. doi: 10.1126/science.1112103. [DOI] [PubMed] [Google Scholar]
- Ono K, Tomasiewicz H, Magnuson T, Rutishauser U. N-CAM mutation inhibits tangential neuronal migration and is phenocopied by enzymatic removal of polysialic acid. Neuron. 1994;13:595–609. doi: 10.1016/0896-6273(94)90028-0. [DOI] [PubMed] [Google Scholar]
- Pencea V, Luskin MB. Prenatal development of the rodent rostral migratory stream. J. Comp. Neurol. 2003;463:402–418. doi: 10.1002/cne.10746. [DOI] [PubMed] [Google Scholar]
- Peretto P, Giachino C, Aimar P, Fasolo A, Bonfanti L. Chain formation and glial tube assembly in the shift from neonatal to adult subventricular zone of the rodent forebrain. J. Comp. Neurol. 2005;487:407–427. doi: 10.1002/cne.20576. [DOI] [PubMed] [Google Scholar]
- Rakic P. Contact regulation of neuronal migration. In: Edelman GM, Thiery JP, editors. The Cell in Contact. Adhesions and Junctions as Morphogenic Determinants. John Wiley; New York: 1985. pp. 67–91. [Google Scholar]
- Rakic P. A century of progress in corticoneurogenesis: from silver impregnation to genetic engineering. Cereb. Cortex. 2006;16(Suppl 1):i3–17. doi: 10.1093/cercor/bhk036. [DOI] [PubMed] [Google Scholar]
- Rougon G, Dubois C, Buckley N, Magnani JL, Zollinger W. A monoclonal antibody against meningococcus group B polysaccharides distinguishes embryonic from adult N-CAM. J. Cell Biol. 1986;103:2429–2437. doi: 10.1083/jcb.103.6.2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousselot P, Lois C, Alvarez-Buylla A. Embryonic (PSA) N-CAM reveals chains of migrating neuroblasts between the lateral ventricle and the olfactory bulb of adult mice. J. Comp. Neurol. 1995;351:51–61. doi: 10.1002/cne.903510106. [DOI] [PubMed] [Google Scholar]
- Sagane K, Hayakawa K, Kai J, Hirohashi T, Takahashi E, Miyamoto N, Ino M, Oki T, Yamazaki K, Nagasu T. Ataxia and peripheral nerve hypomyelination in ADAM22-deficient mice. B.M.C. Neurosci. 2005;6:33. doi: 10.1186/1471-2202-6-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagane K, Ohya Y, Hasegawa Y, Tanaka I. Metalloproteinase-like, disintegrin-like, cysteine-rich proteins MDC2 and MDC3: novel human cellular disintegrins highly expressed in the brain. Biochem. J. 1998;334:93–98. doi: 10.1042/bj3340093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saghatelyan A, de Chevigny A, Schachner M, Lledo PM. Tenascin-R mediates activity-dependent recruitment of neuroblasts in the adult mouse forebrain. Nat. Neurosci. 2004;7:347–356. doi: 10.1038/nn1211. [DOI] [PubMed] [Google Scholar]
- Schwanzel-Fukuda M, Pfaff DW. Origin of luteinizing hormone-releasing hormone neurons. Nature. 1989;338:161–164. doi: 10.1038/338161a0. [DOI] [PubMed] [Google Scholar]
- Stoppini L, Buchs PA, Muller D. A simple method for organotypic cultures of nervous tissue. J. Neurosci. Meth. 1991;37:173–182. doi: 10.1016/0165-0270(91)90128-m. [DOI] [PubMed] [Google Scholar]
- Takahashi E, Sagane K, Oki T, Yamazaki K, Nagasu T, Kuromitsu J. Deficits in spatial learning and motor coordination in ADAM11-deficient mice. B.M.C. Neurosci. 2006;7:19. doi: 10.1186/1471-2202-7-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasiewicz H, Ono K, Yee D, Thompson C, Goridis C, Rutishauser U, Magnuson T. Genetic deletion of a neural cell adhesion molecule variant (N-CAM-180) produces distinct defects in the central nervous system. Neuron. 1993;11:1163–1174. doi: 10.1016/0896-6273(93)90228-j. [DOI] [PubMed] [Google Scholar]
- Tomczuk M, Takahashi Y, Huang J, Murase S, Mistretta M, Klaffky E, Sutherland A, Bolling L, Coonrod S, Marcinkiewicz C, Sheppard D, Stepp MA, White JM. Role of multiple beta 1 integrins in cell adhesion to the disintegrin domains of ADAMs 2 and 3. Exp Cell Res. 2003;290:68–81. doi: 10.1016/s0014-4827(03)00307-0. [DOI] [PubMed] [Google Scholar]
- Tramontin AD, Garcia-Verdugo JM, Lim DA, Alvarez-Buylla A. Postnatal development of radial glia and the ventricular zone (VZ): a continuum of the neural stem cell compartment. Cereb. Cortex. 2003;13:580–587. doi: 10.1093/cercor/13.6.580. [DOI] [PubMed] [Google Scholar]
- Webb DJ, Asmussen H, Murase S, Horwitz AF. Cell migration in slice cultures. Meth. Cell Biol. 2002;69:341–358. doi: 10.1016/s0091-679x(02)69022-3. [DOI] [PubMed] [Google Scholar]
- Weinhold B, Seidenfaden R, Rockle I, Muhlenhoff M, Schertzinger F, Conzelmann S, Marth JD, Gerardy-Schahn R, Hildebrandt H. Genetic ablation of polysialic acid causes severe neurodevelopmental defects rescued by deletion of the neural cell adhesion molecule. J. Biol. Chem. 2005;280:42971–42977. doi: 10.1074/jbc.M511097200. [DOI] [PubMed] [Google Scholar]
- White JM. ADAMs: modulators of cell—cell and cell—matrix interactions. Curr. Opin. Cell Biol. 2003;15:598–606. doi: 10.1016/j.ceb.2003.08.001. [DOI] [PubMed] [Google Scholar]
- Wichterle H, Garcia-Verdugo JM, Alvarez-Buylla A. Direct evidence for homotypic, glia-independent neuronal migration. Neuron. 1997;18:779–791. doi: 10.1016/s0896-6273(00)80317-7. [DOI] [PubMed] [Google Scholar]
- Wolfsberg TG, Bazan JF, Blobel CP, Myles DG, Primakoff P, White JM. The precursor region of a protein active in sperm-egg fusion contains a metalloprotease and a disintegrin domain: structural, functional, and evolutionary implications. Proc. Natl Acad. Sci. USA. 1993;90:10783–10787. doi: 10.1073/pnas.90.22.10783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yachnis AT, Rorke LB. Cerebellar and brainstem development: an overview in relation to Joubert syndrome. J. Child. Neurol. 1999;14:570–573. doi: 10.1177/088307389901400904. [DOI] [PubMed] [Google Scholar]
- Yamashita S, Okada Y. Mechanisms of heat-induced antigen retrieval: analyses in vitro employing SDS-PAGE and immunohistochemistry. J. Histochem. Cytochem. 2005;53:13–21. doi: 10.1177/002215540505300103. [DOI] [PubMed] [Google Scholar]
- Yang P, Baker KA, Hagg T. A disintegrin and metalloprotease 21 (ADAM21) is associated with neurogenesis and axonal growth in developing and adult rodent CNS. J. Comp. Neurol. 2005;490:163–179. doi: 10.1002/cne.20659. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.