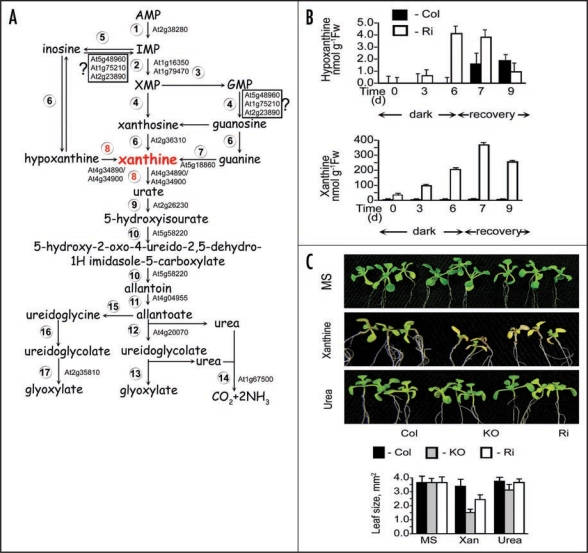
Figure 1.
Purine catabolism, xanthine and hypoxanthine accumulation and Arabidopsis plants growth. (A) Purine nucleotide catabolism in plants. Enzymes shown are: (1) AMP deaminase (EC 3.5.4.6), (2) IMP dehydrogenase (EC 1.1.1.205), (3) GMP synthase (EC 6.3.5.2), (4) 5'-nucleotidase, (5) Nucloeside phosphotransferase (EC 2.7.1.77), (6) Inosine-guanosine nucleosidase (EC 3.2.2.2), (7) Guanine deaminase (EC 3.5.4.15), (8) Xanthine dehydrogenase (EC 1.1.1.204), (9) Uricase EC 1.7.3.3, (10) Hydroxyisourate hydrolase (EC 3.5.2.17),1,18 (11) Allantoinase, allantoin amidohydrolase (EC 3.5.2.5), (12) Allantoicase, allantoate amidohydrolase (EC 3.5.3.4), (13) Ureidoglycolate lyase (EC 4.3.2.3), (14) Urease EC 3.5.1.5, (15) Allantoin deaminase (EC 3.5.3.9), (16) Ureidoglycine amidohydrolase (EC 3.5.3.-), (17) Ureidoglycolate hydrolase (EC 3.5.3.19). (B) Analysis of the purine metabolites, hypoxanthine and xanthine, in response to dark stress. Hypoxanthine and xanthine were determined by HPLC1 in rosette leaves of wild-type (Col) and Ri14, XDH1 RNA interference plants after being kept in dark for 6 days and transferred to a 16-h light/8-h dark regime for recovery over an additional 3 days. Values are means ± SEM (n = 3). (C) Wild-type (Col) and XDH-compromised plants (KO, SALK_148364; Ri, XDH1 RNA interference) were germinated on ¼ MS medium and transplanted on the 5 day to a full MS medium (upper panel) or MS medium with 5.0 mM xanthine and urea as the sole nitrogen source. After transplanting the seedlings were left to grow for 14 days under a 16-h light/8-h dark regime (100 µmol m−2 sec−1) and then photographed. Leaf size was estimated using ImageJ software (http://rsb.info.nih.gov/ij/). Values are means ± SEM (n = 3).