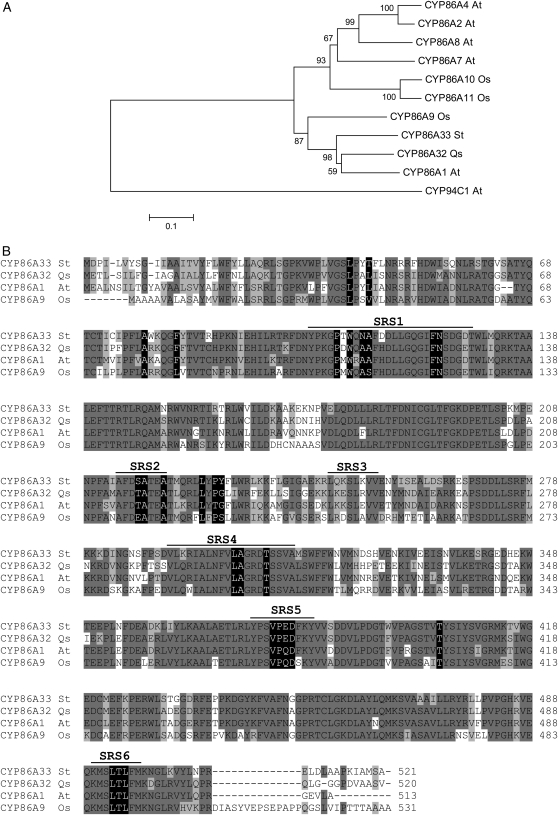
Figure 1.
Phylogenetic analysis of CYP86A subfamily members. A, Phylogenetic analysis of CYP86A32 and CYP86A33. Amino acid sequences of Q. suber (Qs) CYP86A32 and S. tuberosum (St) CYP86A33 were aligned by ClustalW with the CYP86A protein subfamily of Arabidopsis (At) CYP86A1, CYP86A2, CYP86A4, CYP86A7, and CYP86A8 and rice (Oryza sativa; Os) CYP86A9, CYP86A10, and CYP86A11. A CYP94 family member (CYP94C1) was also added. Alignment was used to construct a neighbor-joining tree using the MEGA package, version 3.1. Identical clade distribution and similar bootstrap values were obtained using an unweighted pair group method with arithmetic mean algorithm. Bootstrapped values indicating the level of significance (%) by the separation of the branches are shown. The branch length indicates the extent of the difference according to the scale at the bottom. Note that CYP86A1, CYP86A32, CYP86A33, and CYP86A9 are grouped within the same subclade, although CYP86A9 is separated from the rest. CYP86A and CYP94 are clearly separated in two major clades as previously reported. B, Amino acid sequence alignment of the related proteins CYP86A32, CYP86A33, CYP86A1, and CYP86A9. Amino acids that are identical or similar are shaded in dark or light gray, respectively. SRSs are underlined, and the residues, which it is predicted will contact the oleic acid substrate, are shaded in black.