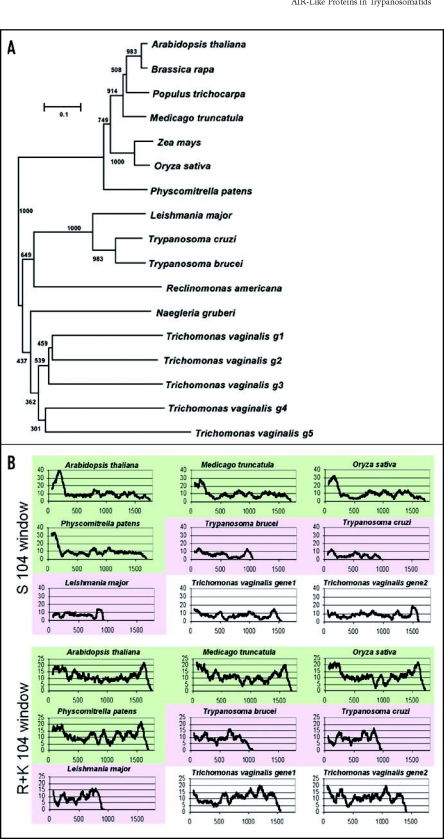
Figure 2.
Phylogenetic relationship of AIR9-like proteins and amino acid scans of plant and protozoan AIR9-like proteins. (A) Phylogenetic comparison based on the LRR domain reveals two subclasses of AIR9-like proteins corresponding to land plants and protozoans, respectively. (B) Amino-acid scans using a 104 amino-acid window. Note the serine-rich regions in AIR9 homologues from land plants all of which present N-terminal peaks above 20 serines in 104 amino acids. otherwise, the sequence similarity between these serine-rich regions is not statistically significant according to BLAST sequence similarity analyses [3]. Arginine plus lysine content in all plant homologues peak above 20 in 104 amino acids in N-terminal (and C-terminal) regions. The N-terminal basic region corresponds to the microtubule-binding domain found in Arabidopsis AIR9. N-terminal basic serine-rich extensions are not found in the AIR9-like proteins from trypanosomatid parasites. Some of the predicted AIR9-like proteins from Trichomonas peak with basic residues in the N-terminus. Green = plant proteins, pink = trypanosomatid proteins, white = Trichomonas proteins. Squences were obtained from protein databases, inferred from EST sequences, or derived from genomic sequences using homologous proteins as template. All sequences are available upon request.