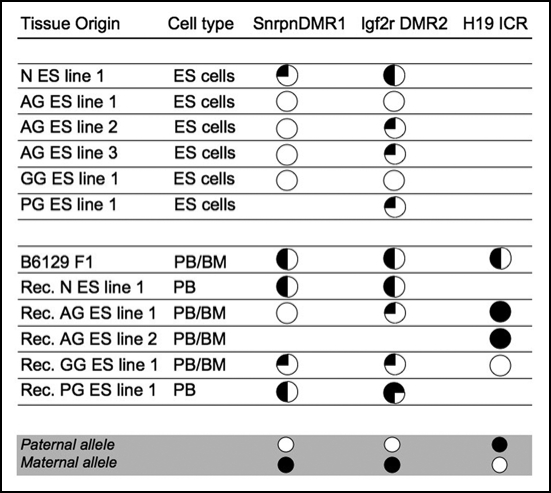
Figure 4.
Methylation of regulatory regions of imprinted genes in uniparental ES cells and their hematopoietic derivatives in recipients. Methylation analysis was performed by bisulfite sequencing.67 Samples were taken from animals with a > 95% contribution (no detectable host or blastocyst component as per Gpi1 analysis) from the ES cell derived component. Circles represent the percentage of methylation detected in all clones (full: methylated, ¼, ½ or ¾: 25, 50 or 75% methylated, empty: not methylated; approximation of methylation to nearest %). Rec., recipient; BM, bone marrow (cell type analyzed for H19); PB, peripheral blood (all other analyses for recipients). The grey bar indicates methylation patterns expected for each parental allele based on analysis of methylation in gametes.101