Abstract
Different classes of biotic (e.g., plant hormones) and abiotic (e.g., different wavelengths of light) signals act through specific signal transduction mechanisms to coordinate all aspects of plant development. Full signal transduction chains have not yet been described for most light or hormonal-mediated events despite the wide range of events early in development which are dependent upon hormonal and light signals. We recently reported a single signal transduction chain which can be initiated by both blue light (BL) and ABA, and which leads to the expression of specific members of the Lhcb gene family in the apical bud of etiolated Arabidopsis seedlings. The signal transduction chain consists of GCR1 (one of two Arabidopsis proteins coding for a potential G-protein coupled receptor), GPA1 (the sole Arabidopsis Ga subunit), PRN1 (Pirin1, one of four members of an iron-containing subgroup of the cupin superfamily), and a Nuclear Factor -Y (NF-Y) heterotrimer comprised of A5, B9 and possibly C9. The same signaling proteins control ABA-mediated delay of germination.
Key Words: blue light, G-protein coupled receptor, G-protein sub unit, abscisic acid (ABA)
Plant Versus Animal Signaling Systems
Heterotrimeric G-protein mediated cell signaling is one of the most highly conserved signaling mechanisms among eukaryotes.1 In terms of signaling components, animals and higher plants have very different genomic situations. Higher plants have one or two copies of G-protein coupled receptors and G protein components, whereas the number present in animal genomes can number in the hundreds.2,3 In Arabidopsis, a GCR1 homolog called GCR2 has recently been reported and characterized as involved in the delay in germination, potentially antagonistic to the role of GCR1.4 The situation is reversed for transcription factors, where plants have numerous types and many copies in a given gene family and animals may have single copy. A good example are the NF-Y factors where the A, B and C subunits in Arabidopsis number ten, ten and nine, respectively,5,6 but in humans there is only one copy of each subunit. This suggests that signaling specificity for plant signal transduction mechanisms may reside at the level of effectors, modifications of signaling components, and/or gene expression itself rather than in the receptors or G-protein sub-types.
The hypothesis presented above is supported by our recent findings that several different effectors are able to relay abiotic or biotic information received from the sole Arabidopsis Gα, in different tissues and at different developmental stages.7 It is also supported by the increasing number of different effector proteins that interact with the Arabidopsis Gα subunit including PRN1,8 Phospholipase C,9 Phospholipase Eα1,10 PD111 and THF1.12
Multiple Plant-Related Signals Share the GCR1-GPA1 System But Not the Same Effectors or Even Further Downstream Components Such as Transcription Factors
The possibility of abiotic and biotic signals stimulating the same pathway is not a new concept in plant biology.11,13–15 Both BL (biotic) and ABA (abiotic) are reported to be able to regulate the same pathway(s) as confirmed by various methodologies.11,16–17 We recently demonstrated using both genetic and photobiological methods, that GCR1 and GPA1, as signal transduction components, are both critical to the ability of BL and ABA to mediate Lhcb expression in etiolated seedlings.7 We also demonstrated that both BL and ABA act through the same signaling chain comprised in part of GCR1 and GPA1, suggesting the possibility that GCR1 can act as a receptor for both BL and for ABA. BL or ABA treatments also result in increased phenylpropanoid production in etiolated seedlings, acting once again via the GCR-GPA1 pathway.11 This is yet further confirmation that the G-protein mediated pathway is core to many signaling pathways and that these pathways diverge at the effector level.
Even beyond the effector level (as outlined previously), in this case PRN1, there is divergence of the pathways depending on tissue type or developmental state of the tissue in which the signaling mechanism is present. NF-Y-B9 (also known as LEC1) is the only “B” subunit with responsible for the BL regulation of Lhcb expression in etiolated Arabidopsis seedlings. However, B9 and B6 (also known as LEC-1-Like) have a role in the mechanism that inhibits the ABA-mediated delay in germination.7,8
Similarly NF-Y-B9 is known to have a solo and critical role in the development of the embryo and events in late embryogenesis, responding to both biotic and abiotic stimuli.18–20 NF-Y-B9 and NF-Y-B6 are both known to interact with proteins found in tissues other than the developing embryo, such as FUS3, itself involved in vegetative processes,21 and ABI3, active in the shoot apex.22–24
The Case for Multifunctional Receptors
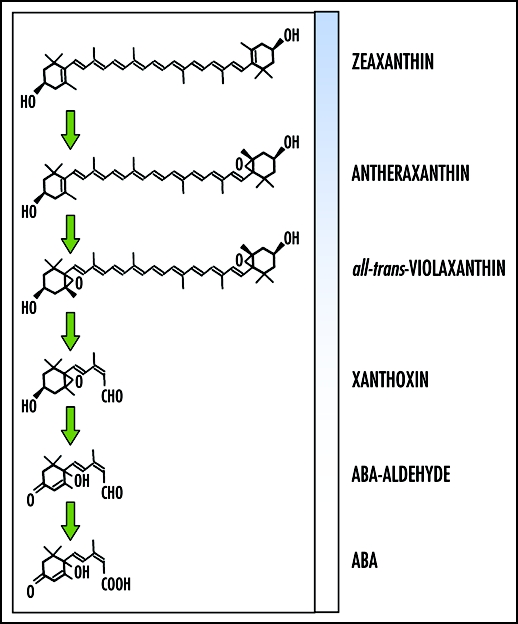
It is unclear how, but clear that, both BL and ABA require GCR1 and use the same proteins of GCR1 and GPA1 in the cotyledons of etiolated seedlings. Light and hormone signals have a different physical nature, so it is reasonable to assume the existence of two separate receptors that might in some way feed into the GCR1-GPA1 system. However, because of the relationship between ABA and BL absorbing carotenoids, we also can propose a “combined” receptor that can respond to both the biotic ABA signal and abiotic BL stimuli. ABA is derived from an asymmetric cleavage of BL-absorbing carotenoids (i.e., zeaxanthin, antheraxanthin, violaxanthin) as shown in Figure 1. Due to the structural relationship between the heterocyclic head group on ABA, and the carotenoid from which it originally derives, it is possible that a single receptor could bind, either ABA or a blue light-absorbing carotenoid.
Figure 1.
ABA synthesis pathway: from BL-absorbing zeaxanthin, to the hormone ABA.
Blue light-absorbing carotenoids are proposed as receptors for some phenomena in plants,25–26 and a carotenoid-based chemical structure is the photoreceptor for rhodopsin, the rod-based photoreceptor which interacts with transducin, a G-protein involved in vision.1 It may be in some systems where there are a paucity of receptors and G-proteins, that over time one receptor has become adopted for use with several signals.
Footnotes
Previously published online as a Plant Signaling & Behavior E-publication: http://www.landesbioscience.com/journals/psb/article/4497
References
- 1.Hamm HE. The many faces of G protein signaling. J Biol Chem. 1998;273:669–672. doi: 10.1074/jbc.273.2.669. [DOI] [PubMed] [Google Scholar]
- 2.Assmann SM. Heterotrimeric and unconventional GTP binding proteins in plant cell signaling. Plant Cell. 2002;14:S355–S373. doi: 10.1105/tpc.001792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perfus-Barbeoch L, Jones AM, Assmann SM. Plant heterotrimeric G protein function: Insights from Arabidopsis and rice mutants. Curr Opin Plant Biol. 2004;7:719–731. doi: 10.1016/j.pbi.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 4.Liu XG, Yue YL, Li B, Nie YL, Li W, Wu WH, Ma LG. A G protein-coupled receptor is a plasma membrane receptor for the plant hormone abscisic acid. Science. 2007;315:1712–1716. doi: 10.1126/science.1135882. [DOI] [PubMed] [Google Scholar]
- 5.Gusmaroli G, Tonelli C, Mantovani R. Regulation of the CCAAT-binding NF-Y subunits in Arabidopsis thaliana. Gene. 2001;264:173–185. doi: 10.1016/s0378-1119(01)00323-7. [DOI] [PubMed] [Google Scholar]
- 6.Gusmaroli G, Tonelli C, Mantovani R. Regulation of novel members of the Arabidopsis thaliana CCAAT-binding nuclear factor Y subunits. Gene. 2002;283:41–48. doi: 10.1016/s0378-1119(01)00833-2. [DOI] [PubMed] [Google Scholar]
- 7.Warpeha KM, Upadhyay S, Yeh J, Adamiak J, Hawkins SI, Lapik YR, Anderson MB, Kaufman LS. The GCR1, GPA1, PRN1, NF-Y signal chain mediates both blue light and ABA responses in Arabidopsis. Plant Physiol. 2007;143:1590–1600. doi: 10.1104/pp.106.089904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lapik Y, Kaufman LS. The Arabidopsis cupin domain protein AtPirin1 and AtGPA1, the Arabidopsis Gα subunit interact with each other and regulate seed germination and early seedling development. Plant Cell. 2003;15:1578–1590. doi: 10.1105/tpc.011890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Apone F, Alyeshmerni N, Wiens K, Chalmers D, Chrispeels MJ, Colucci G. The G-protein-coupled receptor GCR1 regulates DNA synthesis through activation of phosphatidylinositol-specific phospholipase C. Plant Physiol. 2003;133:571–579. doi: 10.1104/pp.103.026005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao J, Wang XM. Arabidopsis phospholipase D alpha 1 interacts with the heterotrimeric G-protein alpha-subunit through a motif analogous to the DRY motif in G-protein-coupled receptors. J Biol Chem. 2004;279:1794–1800. doi: 10.1074/jbc.M309529200. [DOI] [PubMed] [Google Scholar]
- 11.Warpeha KM, Lateef SS, Lapik Y, Anderson MB, Lee BS, Kaufman LS. G-Protein-coupled receptor 1, G-protein Gα-subunit 1, and prephenate dehydratase 1 are required for blue light-induced production of phenylalanine in etiolated Arabidopsis1. Plant Physiol. 2006;140:844–855. doi: 10.1104/pp.105.071282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang J, Taylor JP, Chen JG, Uhrig JF, Schnell DJ, Nakagawa T, Korth K, Jones AM. The plastid protein THYLAKOID FORMATION 1 and the plasma membrane G-Protein GPA1 interact in a novel sugar-signaling mechanism in Arabidopsis. Plant Cell. 2006;18:1226–1238. doi: 10.1105/tpc.105.037259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brocard-Gifford IM, Lynch TJ, Finkelstein RR. Regulatory networks in seeds integrating developmental, abscisic acid, sugar, and light signaling. Plant Physiol. 2003;131:78–92. doi: 10.1104/pp.011916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fan LM, Zhao ZX, Assmann SM. Guard cells: A dynamic signaling model. Curr Opin Plant Biol. 2004;7:537–546. doi: 10.1016/j.pbi.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 15.Zhan X, Wang HB, Takemiya A, Song CP, Kinoshita T, Shimazaki KI. Inhibition of blue light-dependent H+ pumping by abscisic acid through hydrogen peroxide-induced dephosphorylation of the plasma membrane H+-ATPase in guard cell protoplasts. Plant Physiol. 2004;136:4150–4158. doi: 10.1104/pp.104.046573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weatherwax SC, Ong MS, Degenhardt J, Bray EA, Tobin EM. The interaction of light and abscisic acid in the regulation of plant gene expression. Plant Physiol. 1996;111:363–370. doi: 10.1104/pp.111.2.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yadav V, Mallappa C, Gangappa SN, Bhatia S, Chattopadhyay S. A basic helix-loop-helix transcription factor in Arabidopsis, MYC2, acts as a repressor of blue light-mediated photomorphogenic growth. Plant Cell. 2005;17:1953–1966. doi: 10.1105/tpc.105.032060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.West MAL, Yee KM, Danao J, Zimmerman JL, Fischer RL, Goldberg RB, Harada JJ. LEAFY COTYLEDON1 is an essential regulator of late embryogenesis and cotyledon identity in Arabidopsis. Plant Cell. 1994;6:1731–1745. doi: 10.1105/tpc.6.12.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee H, Fischer RL, Goldberg RB, Harada JJ. Arabidopsis LEAFY COTYLEDON1 represents a functionally specialized subunit of the CCAAT binding transcription factor. Proc Natl Acad Sci USA. 2003;100:2152–2156. doi: 10.1073/pnas.0437909100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kwong RW, Bui AQ, Lee H, Kwong LW, Fischer RL, Goldberg RB, Harada JJ. LEAFY COTYLEDON1-LIKE defines a class of regulators essential for embryo development. Plant Cell. 2003;15:5–18. doi: 10.1105/tpc.006973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reidt W, Ellerstrom M, Kolle K, Tewes A, Tiedemann J, Altschmied L, Baumlein H. FUS-3 dependent gene regulation during late embryogenesis. J Plant Physiol. 2001;158:411–418. [Google Scholar]
- 22.Rohde A, De Rycke R, Beeckman T, Engler G, Van Montagu M, Boerjan W. ABI3 affects plastid differentiation in dark-grown Arabidopsis seedlings. Plant Cell. 2000;12:35–52. doi: 10.1105/tpc.12.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rohde A, Van Montagu M, Boerjan W. The ABSCISIC ACID-INSENSITIVE 3 (ABI3) gene is expressed during vegetative quiescence processes in Arabidopsis. Plant Cell Environm. 1999;22:261–270. [Google Scholar]
- 24.Kurup S, Jones HD, Holdsworth MJ. Interactions of the developmental regulator ABI3 with proteins identified from developing Arabidopsis seeds. Plant J. 2000;21:143–156. doi: 10.1046/j.1365-313x.2000.00663.x. [DOI] [PubMed] [Google Scholar]
- 25.Quinones MA, Zeiger E. A putative role of the xanthophyll, zeaxanthin, in blue light photoreception of corn coleoptiles. Science. 1994;264:558–561. doi: 10.1126/science.264.5158.558. [DOI] [PubMed] [Google Scholar]
- 26.Quinones MA, Lu ZM, Zeiger E. Genetic variation of stomatal conductance, blue light sensitivity and zeaxanthin content in guard cells of Pima cotton (Gossypium barbadense) Physiol Plant. 1998;103:560–566. [Google Scholar]