Figure 6.
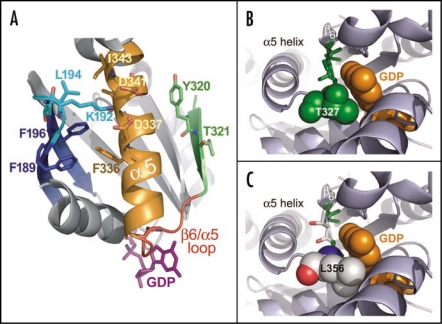
Structural analyses of the α5 helix and the β6/α5 loop in the regulation of GDP release. (A) The β6/α5 loop (red) plays a key role in regulating the binding and release of GDP (magenta sticks). The β6 strand (green) and α5 helix (orange) are each thought to regulate the disposition of this loop. Additionally, the β2/β3 loop (cyan) connecting the β2 and β3 strands (blue) is thought to indirectly affect GDP release by stabilizing the conformation of the α5 helix. Several residues governing this interaction in the β2/β3 loop and α5 helix are shown as sticks. Residues Y320 and T321 in the β6 strand may also stabilize the basal conformation of the α5 helix. Receptor-mediated disruption of these regions may ultimately induce a conformational change in the β6/α5 loop resulting in the release of GDP (reviewed in ref. 82). The structural representation was generated from PDB file: 1BOF.83 (B) Structural representation of residue T327 in Gαi1 highlights the role of β6/α5 loop in nucleotide exchange. Structure of wild type Gαi1 (PDB code: 1BOF)83 illustrates the proximity of the β6/α5 loop to the bound molecule of GDP (orange spheres). The highly conserved TCAT motif (green sticks) within the β6/α5 loop positions the T327 side chain (spheres) directly towards the GDP purine ring. This side chain perfectly accommodates the bound GDP molecule, making several potentially stabilizing contacts (not depicted). (C) AtGPA1 contains a unique TTAL motif within its β6/α5 loop. A modeled mutation of the TCAT motif within the Gαi1 structure to TTAL as present in AtGPA1 (green and white sticks) shows that the leucine side chain (spheres; numbered as in AtGPA1) introduces a likely steric clash with the GDP molecule. This unproductive orientation of the β6/α5 loop is predicted to reduce the affinity for GDP binding and result in a faster nucleotide exchange rate for AtGPA1. The AtGPA1 structural model was generated using the ‘mutagenesis’ function in PyMol (Delano Scientific; San Carlo, CA, USA). The structure of Gαi1 (PDB id 1BOF) was used as the basis for this model and the indicated residues were mutated in silico to corresponding residues in AtGPA1.