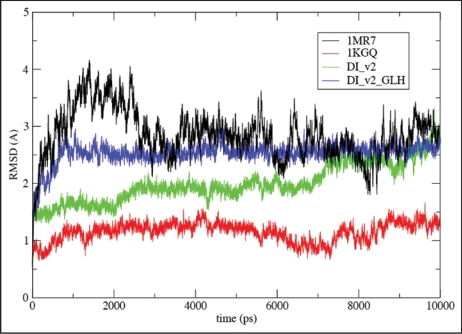
Figure 7.
Root-mean square deviations (RMSD) vs. all atom AMBER8 molecular dynamics64 simulation time to 10 ns from starting structures for LHBH regions of two known proteins (1MR7—Streptogramin A Acetyltransferase68 and 1KGQ—Tetrahydrodipicolinate N-Succinyltransferase69—and two model LHBHs (C4D—diglycoslyated and C4D-D(0)—diglycosylated and protonated glutamate), as shown in Figure 6. In runs up to 1 ns which include five other known LHBH structures we obtain similar RMSDs all bracketed by 1MR7 and 1KGQ. Up to 1 ns, we find similar results for C4M1,C4M2, and N2 (not shown-order 2.5 Å), while the N3 RMSD is much larger (≈6 Å).