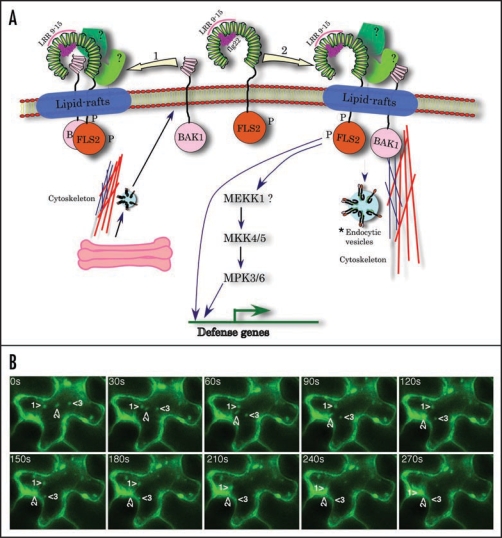
Figure 1.
(A) A hypothetical model for the activation of FLS2. FLS2, after binding flagellin flg22, associates with BAK1 either directly (shown by leftward arrow) or indirectly through other unknown proteins (rightward arrow). In either case the FLS2 complex most likely localizes to lipid-rafts, which include proteins involved in trafficking, signalling and proteolysis. Recent proteomic analysis has shown the presence of BAK1 in plant lipid rafts.34 Engagement of flagellin by FLS2, most likely by its LRR9 to 15,8,9 results in endocytosis of the FLS2 complex either for recycling and/or signalling.20 (A) ‘*’ next to the endocytic vesicle indicates the potential involvement of internalized receptors in activating FLS2 signaling. Evidence for such a mechanism has recently been provided for BRI1 by Geldner et al, (2007).35 The involvement of MEKK1 in FLS2 signaling was provided by Asai et al, (2002).5 However, studies with a mekk1 knockout mutant suggested that MEKK1 is not required for the activation of MPK3 or MPK6 but may play a structural role in FLS2 signaling.38,39 FLS2 may transmit signals independent of endocytosis by activating the MAPK cascade.18,20 Some FLS2-trigged responses may occur independent of MAPK pathway (Prasad et al., unpublished results). The horse-shoe shaped extracellular region of FLS2, which consists of 28 LRR, is derived from the 3D structure of mammalian TLR3. The binding of flagellin to LRR9 to 15 is based on a report by Dunning et al, (2007)9 whereas placing of flg22 on the concave side of LRR is based on mutagenesis and modeling of FLS2 and human TLR5 structure using the known crystal structures of other LRR proteins.9,10 FLS2, FLAGELLIN SENSITIVE2; BAK1, BRI1-ASSOCIATED KINASE1; LRR, leucine rich repeats. (B) FLS2-YFP undergoes exocytosis. FLS2-YFP was expressed in tobacco leaf epidermal cells using the Agrobacterium-mediated transient transformation. Shown are selected images from a time-lapse movie that demonstrate the fusion of several vesicles (marked by numbered arrowheads) with the plasma membrane.