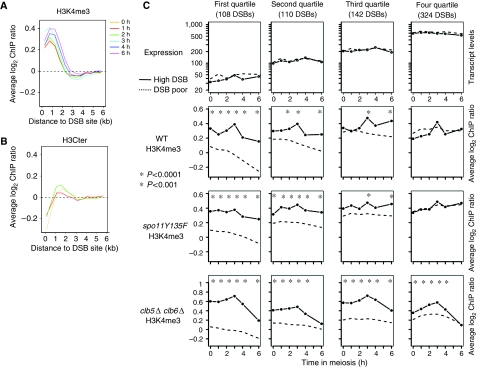
Figure 4.
DSB sites are constitutively hypertrimethylated, independently of transcript levels. (A) Average H3K4me3 as a function of the distance from the DSB sites. All the 1013 DSB hot spot sites were aligned and probes were grouped in bins of 0.5 kb. Average values for each bin are plotted for each indicated time point. (B) Average total H3 levels as a function of the distance from the DSB sites. Same legend as in (A). (C) The non-regulated genes were divided into four quartiles according to their average transcript levels during meiosis. Then DSB sites were examined in each category, both for expression levels of the adjacent genes (top panel), and for H3K4me3 (bottom panels). Profiles of control DSB-poor regions were also calculated for each quartile (dotted lines), as described in Supplementary data. Stars indicate a significant difference between DSB-high and DSB-poor regions. WT, ORD7339; spo11Y135F, ORD7341; clb5Δ clb6Δ, ORD6830.