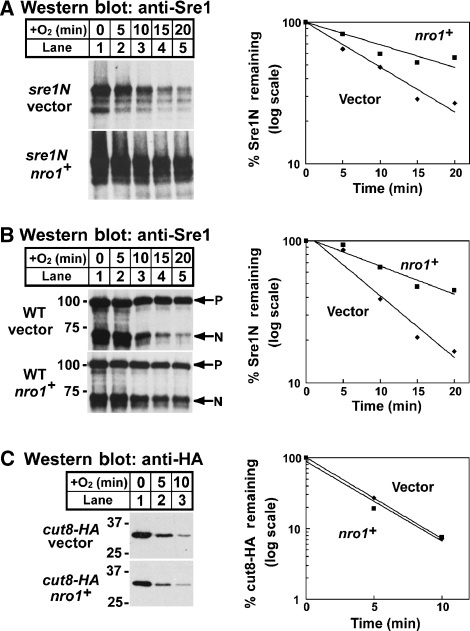
Figure 3.
Nro1 regulates Sre1N degradation. (A) sre1N cells containing empty vector or a plasmid expressing nro1+ were grown in the absence of oxygen for 3 h. At t=0, cycloheximide (200 μg/ml) was added and cells were shifted to the presence of oxygen. (B) Wild-type cells containing empty vector or a plasmid expressing nro1+ were grown in the absence of oxygen for 3 h. At t=0, cycloheximide (200 μg/ml) was added and cells were shifted to the presence of oxygen. P and N denote precursor and N–terminus, respectively. (C) cut8-HA cells containing empty vector or a plasmid expressing nro1+ were grown in the absence of oxygen for 3 h. At t=0, cycloheximide (200 μg/ml) was added and cells were shifted to the presence of oxygen. For each experiment, samples were collected at the indicated times. Whole-cell extracts (40 μg) were subjected to western blot analysis using anti-Sre1 IgG or anti-HA IgG as indicated. The percentage of Sre1N or Cut8-HA remaining at each time point relative to t=0 is quantified to the right; vector (diamonds) and nro1+ (squares). Data shown are representative of four experiments.