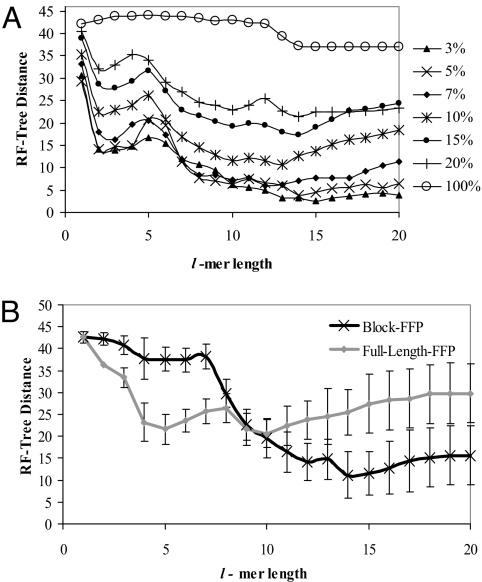
Fig. 3.
Validation of the optimal feature length range by simulation. (A) Tree reconstruction, using FFP comparison of a divergent sequence population. Ten populations of 25 sequences each with a known lineage (the reference tree) were generated with the shuffle model. The substitution rate was varied in 7 trials. UPGMA tree reconstructions were compared with the reference tree, using the Robinson–Foulds (RF) measure. The significance of the peak near l = 4–5 is not known. (B) Tree reconstruction of different length sequences, full-length-FFP vs. block-FFP comparison. 10 trees of 25 sequences were generated using the excision model. The error bars indicate the standard deviation of the 10 trees. The block-FFP method outperforms the full-length-FFP comparison for l ≥ 11. The block length, m = 16,000 is the length of the smallest genome.