Figure 2.
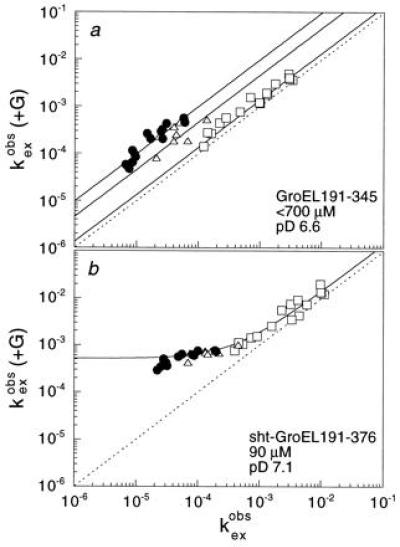
Catalysis of amide proton exchange of barnase
(2.4 mM) by the fragments GroEL191-345 (a) and
sht-GroEL191-376 (b). The results are very similar to
those described in refs. 10 and 11 for intact GroEL. The rate constants
(in units of min−1) for the exchange of individual NH
protons in barnase in the presence of fragment
[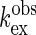 (+G)] are plotted against
those in the absence (
(+G)] are plotted against
those in the absence (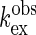 ). Amide
protons that exchange by global, mixed, and local unfolding
mechanisms are displayed by circles, triangles, and squares,
respectively. The plot for 90 μM sht-GroEL191-345 at pD 7.1 (not
shown) is virtually superimposable on that for sht-GroEL191-376
(b). (pD = pH measured in 2H2O.)
It is clearly seen that those protons that require global unfolding for
exchange have significantly increased rates, thus showing that the
fragments bind to the unfolded state of barnase and catalyze its
unfolding. We could only estimate the final concentration of GroEL
fragment in a since GroEL191-345 tended to crystallize
during the exchange experiment at the high initial protein
concentration.
). Amide
protons that exchange by global, mixed, and local unfolding
mechanisms are displayed by circles, triangles, and squares,
respectively. The plot for 90 μM sht-GroEL191-345 at pD 7.1 (not
shown) is virtually superimposable on that for sht-GroEL191-376
(b). (pD = pH measured in 2H2O.)
It is clearly seen that those protons that require global unfolding for
exchange have significantly increased rates, thus showing that the
fragments bind to the unfolded state of barnase and catalyze its
unfolding. We could only estimate the final concentration of GroEL
fragment in a since GroEL191-345 tended to crystallize
during the exchange experiment at the high initial protein
concentration.
