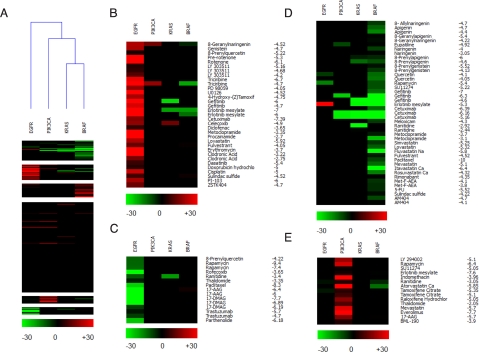
Fig. 3.
Pharmarray analysis of hTERT-HME1 cells carrying the indicated alleles. (A) Heatmap of the pharmacogenomic data (Pharmarray). Each column represents the average of multiple isogenic clones of the indicated genotype. Each row displays the results of differential response to drugs of the KI compared to WT cells. A color code is attributed at each drug concentration tested (log [M]). Drugs that, at the indicated concentrations, preferentially inhibit the growth of mutated cells are highlighted by the red color, while green indicates compounds to which KI cells are more resistant than the WT counterpart. For this reason, the same compound, depending upon its concentration, may segregate in a red (sensitive) or green (resistant) cluster. Black boxes indicate doses at which no significant differences in response between KI and parental cells were observed. Overall clustering of all of the compounds by K-means and of all of the genotypes by hierarchical clustering use an average cosine correlation coefficient. (B–E) Individual clusters composed of drugs with similar genotype-specific activity: (B) EGFR sensitive; (C) EGFR resistant; (D) KRAS/BRAF resistant; (E) PIK3CA-sensitive cluster.