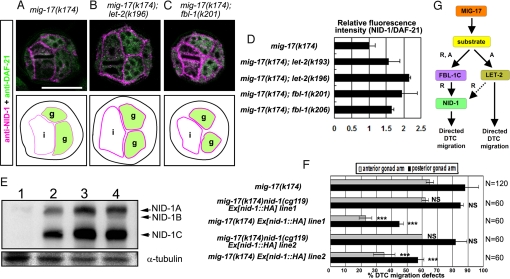
Fig. 5.
NID-1 overexpression suppresses mig-17. (A–C) Cross-sections of L4 hermaphrodites stained with anti-NID-1 (magenta) and anti-DAF-21 (HSP90) (green). DAF-21 was used as an internal standard for quantification of NID-1. (A) mig-17, (B) mig-17; let-2(k196), (C) mig-17; fbl-1(k201). Borders of gonads and intestines are illustrated below the photos. Levels of NID-1 localization to the BM are indicated by magenta thick lines (normal levels) and thin lines (low levels). g: gonads, i: intestines. (Scale bar, 25 μm.) (D) Fluorescence intensity of anti-NID-1 relative to that of anti-DAF-21. The relative fluorescence intensity for each sample was normalized to that of mig-17. See Fig. S6E legend for methods. (E) Extracts from nid-1(cg119) (lane 1), mig-17 (k174) (lane 2), mig-17 Ex[nid-1::HA] line1 (lane 3) or mig-17 Ex[nid-1::HA] line2 (lane 4) hermaphrodites were analyzed. nid-1 expresses three spliced isoforms, NID-1A, B, and C (arrows). Expression of NID-1 proteins are 2.5-fold (line 3) and 2.8-fold (line 4) higher in these transgenic animals compared to mig-17 animals (n = 3). (F) Quantification of DTC migration defects in mig-17 and mig-17 Ex[nid-1::HA] (a NID-1-HA overexpression strain). Results for Fisher's exact test against mig-17(k174) are indicated: ***, P < 0.001; NS, not significant. Error bars represent the mean SD. (G) Model for the protein cascade downstream of MIG-17. MIG-17-dependent proteolysis of an unknown substrate recruits and activates FBL-1C, which then recruits NID-1 to the BM to control DTC migration. MIG-17-dependent proteolysis also activates LET-2 to induce NID-1-dependent and -independent MIG-17 pathways. Dashed arrow, potentially indirect interaction. R, recruitment; A, activation.