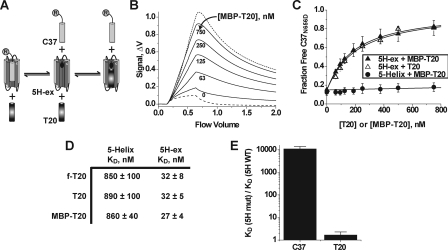
FIGURE 3.
Properties of the 5H-ex/T20 interaction. A, overview of the C37 displacement assay. T20 and R-C37 compete for the same binding site on 5H-ex, and therefore titration of T20 (or MBP-T20) leads to an increase in the free (unbound) concentration of R-C37. The circled R denotes the rhodamine label linked to C37. B, Kinexa 3000 fluorescence response as solutions of 5H-ex (10 nm), R-C37N656D (1 nm), and MBP-T20 (indicated concentration) were passed over beads that specifically captured unbound R-C37N656D. The dashed and dotted lines were generated by control samples in which R-C37N656D is fully bound or fully unbound, respectively (see “Experimental Procedures”). The fraction of unbound R-C37N656D in each sample was calculated by normalizing the difference fluorescence signal Δf to the minimum and maximum Δf obtained from the control solutions. C, C37N656D displacement from 5H-ex (triangles) or 5-Helix (circles) as T20 (open symbols) or MBP-T20 (filled symbols) was titrated into reaction mixtures. The data points represent the means ± S.E. of at least three separate measurements and have been fit to a three-state competitive binding model (solid lines). D, tabulated KD values for the interaction of T20 with either 5-Helix or 5H-ex as determined using the direct binding assay (f-T20) or the C37 displacement assay (T20, MBP-T20). The values represent the means and S.E. of three or more separate measurements. E, effect of the W571R/G572D substitution in the N-HR hydrophobic pocket on C-peptide binding affinities. The data are represented as fold change in KD value relative to the wild type 5H-ex/C-peptide KD. The KD values were determined using the direct binding assay for C37 values or the C37 displacement assays for MBP-T20 values.