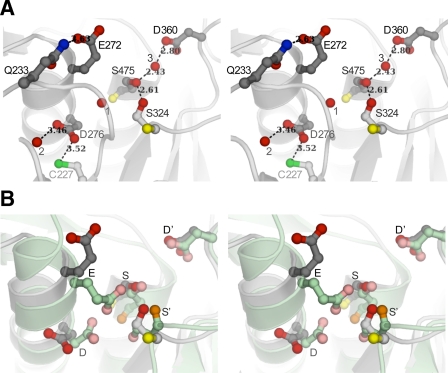
FIGURE 4.
Comparison of the active site of pro-TPP1 zymogen with that of mature kumamolisin. Backbones are shown as ribbon representations with featured side chains in ball-and-stick mode, bound water molecules depicted as red spheres, and hydrogen bonds shown as dashed lines with distances in Å. A, stereo view of TPP1 showing catalytic residues in a conformation that is recalcitrant to activity. In TPP1, Glu272 (E), Asp276 (D), and Ser475 (S) form the catalytic triad, and Asp360 (D′) forms part of the stabilizing oxyanion hole. Ser324 (S′) is completely conserved in the S53 family. Backbone carbonyl groups are shown only for the Ser and S′ residues. Side chain oxygen atoms are shown in red and backbone carbonyl oxygens in yellow. Both Glu272 and Asp276 side chains are oriented away from Ser475. The position of Glu272 is stabilized by a hydrogen bond to Gln233. A bound water molecule (1) occupies the position expected for one of the carboxyl oxygens of Glu272, if the active site were poised for activity. One of the carboxyl oxygens of Asp276 forms a hydrogen bond to a water molecule (2), and the other is within weak hydrogen bond distance to the sulfhydryl of unpaired Cys277. The side chain hydroxyls of Ser475 and Ser324 participate in a strong hydrogen bond. A third water molecule (3) makes a strong hydrogen bond with the Ser475 hydroxyl and is also within hydrogen bond distance of the Asp360 carboxyl constituting the oxyanion hole, close to where the carbonyl oxygen of the scissile peptide bond is expected to be positioned. B, comparison of catalytic machinery of pro-TPP1 with that in mature kumamolisin. Least square superpositions of Cα coordinates of the pro-TPP1 catalytic domain with those of all available structures of S53 family members were performed and yielded similar results. Superposition of TPP1 with mature kumamolisin (PDB ID 1GTL) (27) is shown with ribbon depictions in gray and green, respectively. Although the corresponding oxyanion D′ residues in TPP1 and kumamolisin are in similar conformations, the corresponding catalytic Glu and Asp residues are both in different conformations. Furthermore, the S′ residue in TPP1 (Ser324) is in a different rotameric conformation, and its Cα position is displaced by 1.5 Å from the corresponding S′ (Ser316) in kumamolisin, resulting in different hydrogen bond schemes with the active site Ser.