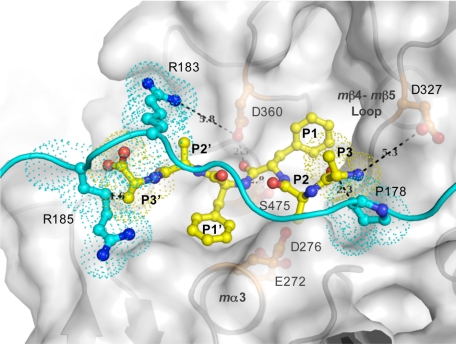
FIGURE 5.
Model of peptide substrate in the TPP1 active site. The model was constructed by docking a hexapeptide substrate, AAFFAA, with a fixed position of the carbonyl oxygen of the P1 residue (Phe) relative to the catalytic domain of TPP1, based on the prokumamolisin structure (PDB ID 1T1E) (28), onto the catalytic domain of TPP1 in which the side chains of active site residues Glu272 and Asp276 were reoriented in the presumed active conformation as found in mature bacterial homologs. A ribbon diagram and a transparent surface rendition of the catalytic domain are shown in gray, a ball-and-stick depiction of the modeled hexapeptide substrate in yellow, and the superimposed linker with side chains of featured residues shown in ball-and-stick mode in cyan. Overlapping residues are shown in dotted surface. This model is an approximation because the positions of the mα3 helix with Glu272 and Asp276 as well as mβ4 with Ser324 together with the mβ4-mβ5 loop with Asp327 are likely to differ in the mature enzyme (Fig. 4). The N terminus of the tripeptide substrate is likely to interact with the Asp327 side chain carboxyl groups. The predicted protonation state of Asp327 (see text) is consistent with a role in binding to an unsubstituted positively charged N terminus of the P3 residue of a putative substrate, thus providing a possible structural basis for the tripeptidyl peptidase activity. In addition, in the absence of crystal lattice contacts, positively charged Arg183 of the linker is likely to participate in electrostatic interactions with negatively charged Asp360 of the oxyanion hole, potentially restricting its role in general acid and base catalysis.