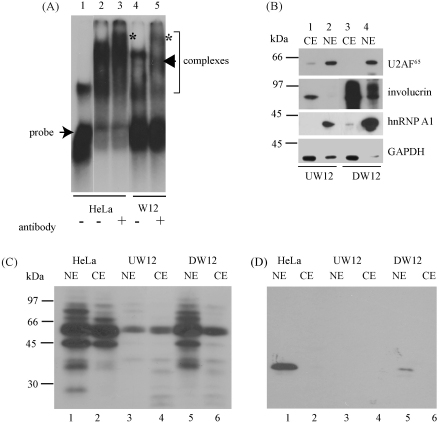
Fig. 4.
hnRNP A1 binds the HPV16 LRE. (A) Electrophoretic mobility shift assay showing interaction of hnRNP A1 and LRE RNA in HeLa and W12 cells. Free probe and the RNA/protein complexes formed are indicated. Asterisks and an arrowhead indicate RNA/protein complexes that have been supershifted due to binding of the anti-hnRNP A1 antibody in tracks 3 and 5. Track 1, [α-32P] rUTP-labelled LRE RNA probe alone (the LRE RNA is a stem loop structure (Cumming et al., 2003); the upper band is due to secondary structure formation); track 2, LRE RNA probe incubated with HeLa nuclear extract; track 3, as in track 2 but nuclear extract preincubated with anti-hnRNP A1 antibody; track 4, LRE RNA probe incubated with W12E nuclear extract; track 5, as in track 4 but nuclear extract preincubated with anti-hnRNP A1 antibody. (B) Western blot of nuclear (tracks 2 and 4) and cytoplasmic (tracks 1 and 3) extracts from undifferentiated (tracks 1 and 2) and differentiated (tracks 3 and 4) W12E cells showing fractionation of U2AF65 and hnRNP A1 mainly with the nuclear fraction and involucrin mainly with the cytoplasmic fraction as expected. The increase in levels of involucrin in tracks 3 and 4 confirms that the W12 cells have differentiated. (C) Autoradiogram of an SDS-PAGE fractionation of UV crosslinking of [α-32P] rUTP-labelled LRE RNA probe with nuclear (NE) and cytoplasmic (CE) fractions of HeLa and undifferentiated (U) and differentiated (D) W12E cells. (D) Western blot of the SDS-PAGE in (C) probed with anti-hnRNP A1 antibody.