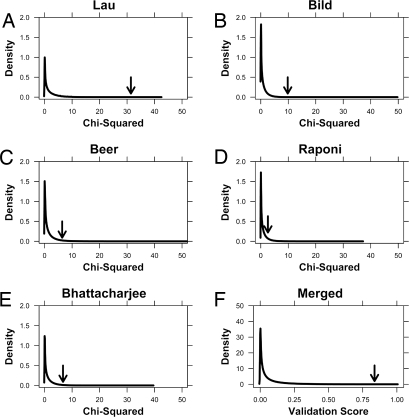
Fig. 3.
Permutation validation. Ten million 6-gene signatures were generated at random from our training dataset. The ability of each signature to separate the training dataset into 2 groups with significantly different prognoses was evaluated using the log-rank test. The kernel density of the χ2 values from this log-rank test was generated (A). The x axis indicates the χ2 values: Larger values indicate a lower P value and hence a more statistically significant separation of patient groups in the training dataset. The y axis gives the kernel density, which reflects the probability distribution of the dataset. Higher values indicate a larger fraction of the population, akin to a smoothed histogram. The performance of the mSD signature is marked with an arrow. These 10 million trained signatures were then tested in 4 independent datasets. Kernel density estimates, as above, are provided for each test dataset (B–E). Each test dataset is labeled with the first author of the study. The performance of the mSD signature is marked with an arrow. Finally, to demonstrate the significance of the mSD signature across all 4 test datasets we generated a validation score by multiplying the percentile rankings of each signature in each of the 4 test datasets. Higher values thus correspond to improved validation across all 4 datasets. The performance of the mSD signature is marked with an arrow.