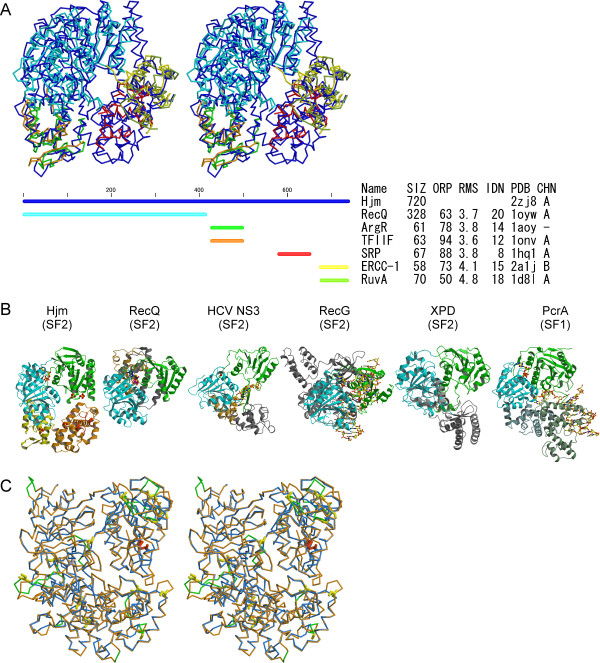
Figure 4.
Schematic view of the structural alignment. (A) Domain structure conservation of PfuHjm with relevant DNA binding proteins. SIZ, ORP, RMS, and IDN are the number of residues aligned, percent superposed residues (% overlap), RMSD (Å), and sequence identity (%) of each domain to Hjm, respectively. PDB and CHN are the PDB code and chain ID of the solutions, respectively. Abbreviations, Hjm; PfuHjm, RecQ; E. coli RecQ, ArgR; E. coli Arginine repressor, TFIIF; Transcription initiation factor IIF, SRP; single recognition particle protein, ERCC-1; Human DNA endonuclease ERCC-1, RuvA; E. coli Holliday junction binding protein RuvA. (B) Structure comparison of Hjm with other relevant proteins. Structures are aligned using the two well-conserved helicase domains (cyan and green), to highlight the variation of the other domains and those orientation. (C) Superposition of PfuHjm (blue) and the homology model of the human DNA polymerase Θ (orange) helicase domain, shown in stereo figure. The major-insertions (with more than two residues) of the DNA polymerase Θ were colored green. The insertions were localized to the peripherals of the molecule, and the central crafts of the proteins are mostly intact. The seventeen cysteine residues of the DNA polymerase Θ are shown in stick models, and colored according to their possible characteristics (buried, grey; exposed, yellow; disulfide bond, red)