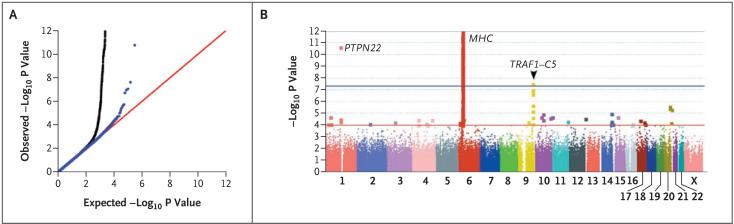
Figure 1. Results of the Genomewide Association Study.
Panels A and B show results for 297,086 polymorphic SNPs genotyped in samples from 1522 case subjects with rheumatoid arthritis who were seropositive for autoantibodies against cyclic citrullinated peptide (anti-CCP-positive) and 1850 control subjects from the combined data sets from the North American Rheumatoid Arthritis Consortium (NARAC) and the Swedish Epidemiological Investigation of Rheumatoid Arthritis (EIRA). Panel A shows a comparison of distributions of observed versus expected P values generated by Cochran-Mantel-Haenszel stratified analysis and corrected for residual inflation by genomic control. Black data points represent the inclusion of SNPs from the major-histocompatibility-complex (MHC) locus; blue data points represent the exclusion of MHC. The most significant non-MHC SNPs are at PTPN22 and the TRAF1-C5 locus. Panel B shows SNPs plotted according to chromosomal location, with the -log10 P values corrected with the use of genomic control. The blue horizontal line indicates SNPs that are significant at a genomewide level (P = 5×10-8). SNPs at PTPN22 and within the MHC locus, where the HLA-DRB1 gene resides, reach genomewide significance. Multiple SNPs across the TRAF1-C5 locus on chromosome 9 (e.g., rs3761847; P = 2×10-8 have a significant association.