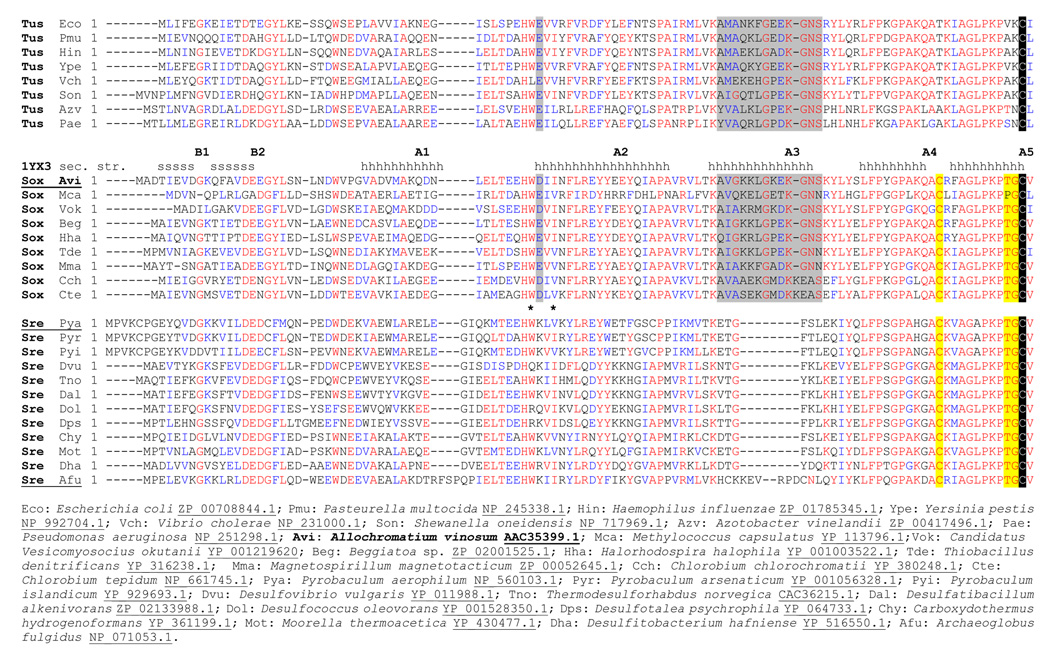
Fig. 1. Multiple sequence alignment of DsrC/TusE sequences.
Sequences are grouped according to whether the host organism’s genome has tusBCD but lacks dsrAB (Tus), has dsrEFH and dsrAB (Sox, sulfur oxidizers), or lacks dsrEFH but has dsrAB (Sre, sulfate or sulfite reducers). These groupings to correspond well to those seen in the phylogenetic tree cladogram, with the exception that DsrC sequences from Pyrobaculuum species were grouped more closely with sequences from dsrEFH-containing-sulfur oxidizers, rather than with sulfate-reducing bacteria. Below the sequence alignment are shown the full organism names and corresponding database accession numbers for each sequence. Positions of secondary structure elements in A. vinosum DsrC are indicated above its sequence. Residue insertions specific to sulfate reducers or TusBCD/DsrEFH-containing organisms are marked with asterisks (*) and grey shading, respectively. C-terminal residues specific to DsrAB-containing sulfur oxidizers or sulfate reducers are identified with yellow shading. The invariant penultimate cysteine is shaded black.