Figure 2.
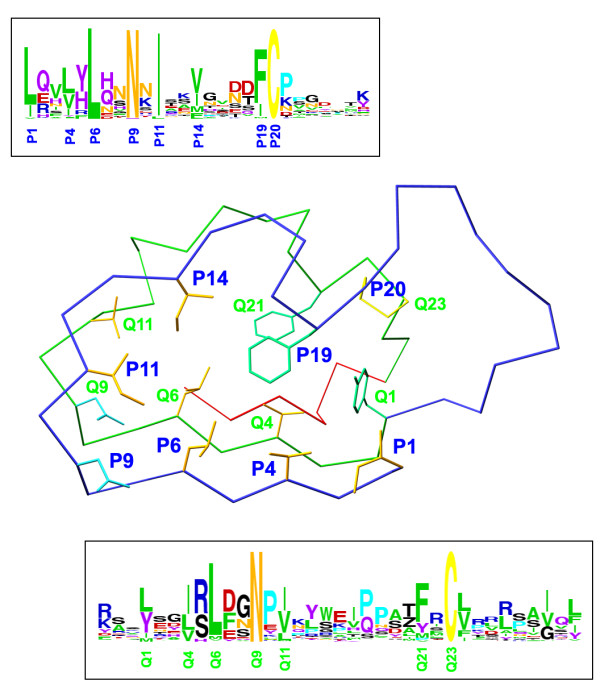
Mapping of the regular expression pattern of the LRRCE motif on a skeletal representation of the LRRCE structure from bovine decorin [29]. The motif includes the laterally extended ear repeat, shown as a Cα trace in blue, the following LRR (in green Cα trace), and the final β-strand closing the domain (red Cα trace). The regular expression pattern used in this study, written in PROSITE syntax [79], was: [LIV]-X(2)-[LVIYFMA]-X-[LIFM]-X(2)-[NH]-X-[ILVF]-X(2)-[VIMFLY]-X(4)-[FIMLV]-C-X(7,20)-[LYIMV]-X(2)-[ILVTMF]-X-[LVMI]-X(2)-N-X-[IVLMAFT]-X(8,9)-[FYMPVAIS]-X-C. In PROSITE syntax each conserved position is shown either as a single amino acid (e.g. C, N) or all possible amino acids for that position enclosed within brackets (e.g. [ILVF] indicates that such position is occupied by Ile, Leu, Val or Phe); each variable position is shown with a letter X. Numbers in parentheses indicate stretches of variable positions (e.g. X(7,20) indicates a stretch of between 7 and 20 variable amino acids). Amino acid preferences for each position are shown in two boxes in "weblogo" form [87]. The conserved sequence positions for the ear repeat on the LRRCE motif are designated as P1, P4, P6,..., P20, and those for the following LRR as Q1, Q4, Q6,..., Q23. The side chains show the amino acids occurring at these conserved positons in the structure of bovine decorin.
