EMBO J 28, 193–204 (2009); published online 4 February 2009
Three different laboratories have now identified a new morphogenetic factor widely conserved in bacteria. The protein, RodZ, is required for assembly of the actin cytoskeleton MreB that controls cell wall synthesis and cell shape.
It is not yet understood how bacteria determine their shape. Almost all bacteria are surrounded by a giant cell wall polymer called peptidoglycan that gives shape to the cells and is an important target of antibiotics. Thus, one main aim is to understand the highly complex enzymatic machinery that synthesizes peptidoglycan and how its activity is coordinated with cell growth and division. The rod-shaped bacteria Escherichia coli and Bacillus subtilis and the curved Caulobacter crescentus have been used extensively as models in the study of cell morphogenesis. A paradigm has emerged in which the essential actin homologue MreB, discovered by Masaaki Wachi many years ago (Wachi et al, 1987), is a key player. In the beginning of the decade, it was shown that MreB of B. subtilis forms helical structures beneath the cell surface and that these structures are required to maintain cell shape (Jones et al, 2001). Later studies confirmed that MreBs of E. coli and C. crescentus (Kruse et al, 2003; Figge et al, 2004) played a similar role. MreB interacts with MreC and MreD and these latter proteins are also essential to cell shape maintenance (Kruse et al, 2005). MreB and MreC are inner membrane proteins much less abundant than MreB (Wachi et al, 2006). Bacterial two-hybrid analyses and pull-down experiments showed that MreC interacts with the penicillin-binding proteins that synthesize the cell wall and therefore are cell shape determinants (Divakaruni et al, 2005, 2007; van den Ent et al, 2006). These results raised the possibility that the MreB filaments interact with MreCD complexes located in the inner cell membrane and thereby control the activity of the external cell wall-synthesizing protein complexes in space and time. This conjecture was supported by the observation that MreC forms helical structures that alternate with the MreB helices (Dye et al, 2005). This simple model is shown schematically in Figure 1.
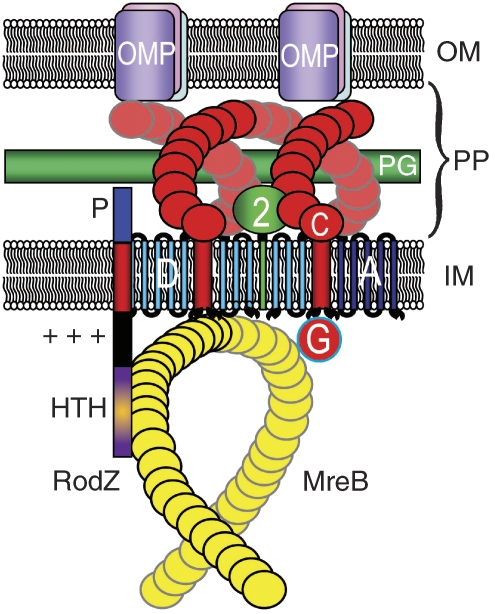
Figure 1.
Schematic diagram that visualizes the interactions between RodZ and the MreBCD–MrdAB (PBP2 RodA) complexes of E. coli. RodZ is shown as a vertical bar with four domains. The HTH domain (magenta) mediates interaction with MreB, whereas the juxta-membrane (JM) domain (+++) is in close contact with the negatively charged membrane phospholipids and serves to configure MreB in its helix-like appearance. P is the periplasmic part of RodZ that interacts with as yet unknown components in the periplasm. MreB is shown as a yellow helix beneath the inside of the inner membrane (IM) that interacts with MreC (red helix). MreC, in turn, interacts with PBP2 (green) and different OMPs. PP, periplasm; OM, outer membrane; PG, peptidoglycan layer; D, MreD in the IM; A, RodA in the IM.
A new common player in bacterial cell morphogenesis has now been discovered independently by three different laboratories, published in separate issues of the EMBO Journal (Shiomi et al, 2008; Bendezu et al, 2009), and Proc Natl Acad Sci USA (Alyahya et al, 2009). Hironori Niki's group screened the Keio strain collection of gene deletions and thereby identified a novel gene, rodZ (yfgA) that is required to maintain proper cell shape. Cells lacking the rodZ gene were round or otherwise misshapen and exhibited a highly reduced growth rate. The diameters of the majority of the rodZ-null cells were similar to that of the width of wild-type E. coli cells and the Niki group therefore suggested that RodZ is a primary determinant of cell length. In that model, cell width is maintained by the MreBCD and PBP2/RodA complexes. Overproduction of RodZ resulted in an increased cell length with little or no change of cell width, consistent with the model.
Piet de Boer's group identified rodZ by screening for the requirement for extra FtsZ, as it was known that increased FtsZ levels rescue the lack of other cell shape determinants (i.e. MreB and PBP2) (Bendezu and de Boer, 2008). They also found that rodZ-null cells exhibited a cold-sensitive phenotype. At low temperatures, the cells were non-dividing large and misshapen spheres.
RodZ is a remarkable multidomain protein that, similar to MreB, forms helical structures associated with the cell membrane (Figure 1). RodZ has one single trans-membrane domain that divides the protein into a cytosolic and a periplasmic part. Interestingly, RodZ has a helix-turn-helix DNA-binding motif (λ Cro type) in its N-terminal cytosolic domain. A deletion analysis showed that the HTH motif was required for the formation of RodZ helices and for full complementation of the defective shape of rodZ-null cells. The function of the HTH domain is unknown but bacterial two-hybrid data indicated that it may mediate important interactions between RodZ and MreB. However, it is also possible that the HTH motif somehow links the nucleoid to the cell wall. The detailed deletion analysis by the de Boer group showed that the basic juxta-membrane (JM) domain of RodZ (marked as +++ in Figure 1) is the only one that is strictly required for the maintenance of the rod shape. The JM domain needs to be membrane associated, and also requires either the HTH domain or the periplasmic domain of RodZ to be functional. These results suggest a scenario in which RodZ interacts with components of the MreBCD–MrdAB (PBP2–RodA) machinery at either side of the membrane and that one of these interactions is sufficient to incorporate the JM domain into the MreB helix and thereby confer cell shape maintenance.
Bendezu et al (2009) constructed a fully functional mreB∷mCherry sandwich fusion and obtained convincing evidence that the formation of MreB and RodZ helices were interdependent, that is, both proteins were required for the helical structures to form. Co-overexpression of MreB and RodZ showed that maintenance of cell shape depended critically on a proper MreB/RodZ ratio. Both groups agreed that MreB and RodZ colocalized in helical structures, but Shiomi et al (2008) suggested that RodZ can form helices independently of MreB. Future work is required to resolve this apparent inconsistency.
Independently of the work of the de Boer and Niki groups, Christine Jacobs-Wagner's group identified RodZ as a morphogenetic determinant of C. crescentus and, importantly, B. subtilis (Alyahya et al, 2009). Supporting the view of Bendezu et al (2009), the subcellular localization pattern of RodZ depended on MreB and furthermore corresponded to active sites of peptidoglycan synthesis. In summary, all three papers identified a new, highly conserved morphogenetic factor and thereby have opened new possibilities in the difficult but essential analysis of the bacterial cell wall puzzle.
Acknowledgments
I thank David M Bulmer for help with Figure 1. This study was supported by a grant from the UK Biotechnology and Biological Sciences Research Council.
References
- Alyahya SA, Alexander R, Costa T, Henriques AO, Emonet T, Jacobs-Wagner C (2009) RodZ, a component of the bacterial core morphogenic apparatus. Proc Natl Acad Sci USA (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendezu F, de Boer PA (2008) Conditional lethality, division defects, membrane involution, and endocytosis in mre and mrd shape mutants of Escherichia coli. J Bacteriol 190: 1792–1811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendezu FO, Hale CA, Bernhardt TG, de Boer P (2009) RodZ (YfgA) is required for proper assembly of the MreB actin cytoskeleton and cell shape in E. coli. EMBO J (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divakaruni AV, Baida C, White CL, Gober JW (2007) The cell shape proteins MreB and MreC control cell morphogenesis by positioning cell wall synthetic complexes. Mol Microbiol 66: 174–188 [DOI] [PubMed] [Google Scholar]
- Divakaruni AV, Loo RR, Xie Y, Loo JA, Gober JW (2005) The cell-shape protein MreC interacts with extracytoplasmic proteins including cell wall assembly complexes in Caulobacter crescentus. Proc Natl Acad Sci USA 102: 18602–18607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dye NA, Pincus Z, Theriot JA, Shapiro L, Gitai Z (2005) Two independent spiral structures control cell shape in Caulobacter. Proc Natl Acad Sci USA 102: 18608–18613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figge RM, Divakaruni AV, Gober JW (2004) MreB, the cell shape-determining bacterial actin homologue, co-ordinates cell wall morphogenesis in Caulobacter crescentus. Mol Microbiol 51: 1321–1332 [DOI] [PubMed] [Google Scholar]
- Jones LJ, Carballido-Lopez R, Errington J (2001) Control of cell shape in bacteria: helical, actin-like filaments in Bacillus subtilis. Cell 104: 913–922 [DOI] [PubMed] [Google Scholar]
- Kruse T, Bork-Jensen J, Gerdes K (2005) The morphogenetic MreBCD proteins of Escherichia coli form an essential membrane-bound complex. Mol Microbiol 55: 78–89 [DOI] [PubMed] [Google Scholar]
- Kruse T, Møller-Jensen J, Løbner-Olesen A, Gerdes K (2003) Dysfunctional MreB inhibits chromosome segregation in Escherichia coli. EMBO J 22: 5283–5292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiomi D, Sakai M, Niki H (2008) Determination of bacterial rod shape by a novel cytoskeletal membrane protein. EMBO J 27: 3081–3091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Ent F, Leaver M, Bendezu F, Errington J, de BP, Lowe J (2006) Dimeric structure of the cell shape protein MreC and its functional implications. Mol Microbiol 62: 1631–1642 [DOI] [PubMed] [Google Scholar]
- Wachi M, Doi M, Tamaki S, Park W, Nakajima-Iijima S, Matsuhashi M (1987) Mutant isolation and molecular cloning of mre genes, which determine cell shape, sensitivity to mecillinam, and amount of penicillin-binding proteins in Escherichia coli. J Bacteriol 169: 4935–4940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachi M, Osaka K, Kohama T, Sasaki K, Ohtsu I, Iwai N, Takada A, Nagai K (2006) Transcriptional analysis of the Escherichia coli mreBCD genes responsible for morphogenesis and chromosome segregation. Biosci Biotechnol Biochem 70: 2712–2719 [DOI] [PubMed] [Google Scholar]