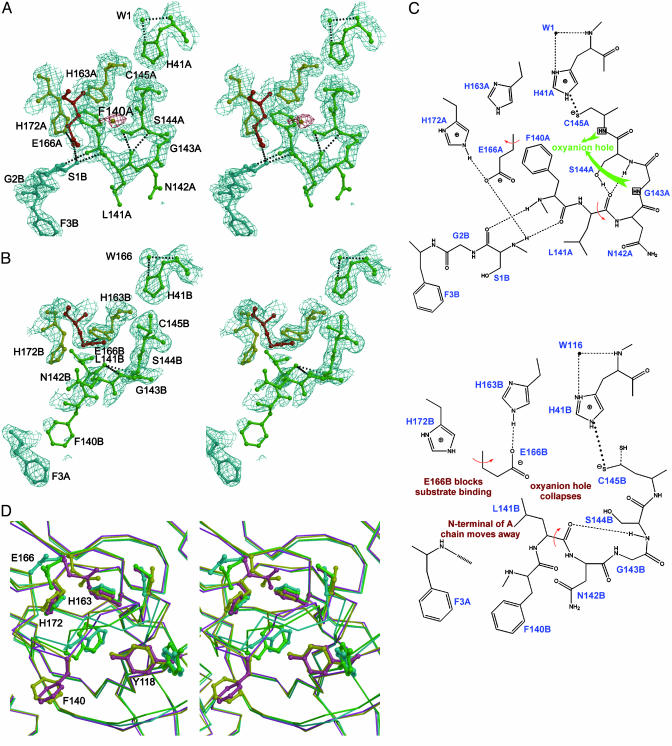
Fig. 2.
Conformational variations in the S1 substrate-binding pocket. (A) A stereoview of the active site of protomer A built into the 1.9-Å electron density (2 Fo - Fc, contoured at 1.0 σ). The oval-shaped piece of electron density, which is red, is assigned to a water molecule. In S1 subsite of protomer A, Glu-A166 is red, His-A163 and His-A172 are yellow, and the other residues are green. Protomer B is cyan. The amino acid residues of the protein are labeled in single letters; for example, H163A stands for His-163 of monomer A (i.e., His-A163). (B) A stereo image showing the collapsed active site of protomer B built into electron density (2Fo - Fc, contoured at 1.0 σ). The oxyanion hole collapses, the N-finger of chain A is not anchored to its binding site on protomer B, Phe-B140 is directed out into bulk solvent, and Glu-B166 switches conformation to block the substrate-binding site. (C) A schematic presentation of the conformational variations and altered hydrogen-bonding networks in active sites. (Upper) The oxyanion hole (for protomer A) and N-finger of protomer B docked to its binding site. (Lower) The corresponding view of the collapsed active site in protomer B. The N-finger is not docked to its binding site, with the following consequences: (i) the oxyanion hole collapses; (ii) Phe-B140 protrudes into bulk solvent; and (iii) Glu-B166 switches conformation to block the S1 substrate-binding subsite. (D) Comparison of four SARS-CoV Mpro structures. A stereo figure is shown of the substrate-binding pocket of protomer B, with their Cα superimposed. The coloring is as follows: pH 6.0, yellow; pH 7.6, cyan; pH 8.0, green; and CMK inhibitor complex, pink. Side chains are shown as ball-and-stick models for the residues Tyr-B118, Phe-B140, Cys-B145, His-B163, Glu-B166, and His-B172. Note the dramatic conformational changes for Tyr-B118, Phe-B140, Cys-B145, and Glu-B166 when the pH changes from 6.0 to higher pH values.