Figure 3.
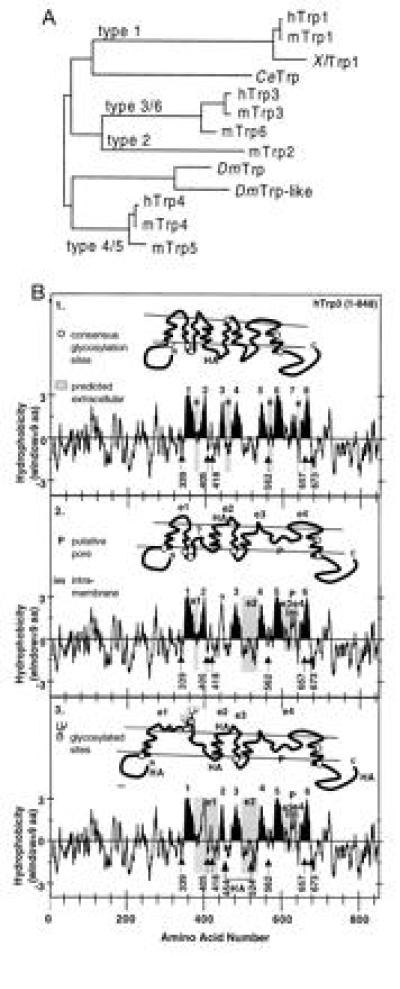
Molecular diversity of 12 Trp sequences as deduced from their corresponding cDNAs and early speculations on secondary structure of CCE channels. (A) Phylogenetic tree obtained by pair-wise comparison of the amino acid sequences that span the putative pore region and the two putative transmembrane segments that flank this region (≈170 amino acids). This assignment assumes a transmembrane topology homologous to that of voltage-gated Ca2+, Na+, and K+ channels (B2 and 3). (B) Kyte–Doolittle plots and models of transmembrane topology of hTrp3 highlighting in black eight or two versions with six hydrophobic regions that may form transmembrane segments based on a possible homologous relation to voltage-gated ion channels. The location of consensus NXS/T glycosylation sites are highlighted (numbers correspond to the asparagines of the NXS/T motifs). In the six-transmembrane models, one of the hydrophobic regions thought not to span the membrane completely, P, is believed to be intramembrane (im) and to form the putative SS1-2 pore region used for the construction of the phylogenetic tree shown in A. e and e1-e4 are sequences predicted by the various models to be exposed to the outside of the cell. Note that, in B2, none of the six consensus glycosylation sites (○) is predicted to be glycosylated, while, in B3, two sites are exposed and presumed to be glycosylated. Locations at which HA epitopes were introduced are also shown.
