Table 2. Sites under PDS in the A. thaliana MADS-box gene family: “Branch-site analyses”.
| Branch-site analyses | n |
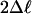 M3 (K = 2) vs. MB (df 2) M3 (K = 2) vs. MB (df 2) |
Parameter estimates under MB | Positively selected sites under MB |
|---|---|---|---|---|
| Type II | ||||
| Branch leading to the FLC-like genes | 39 | 20.50*** | P0 = 0.45, P1 = 0.38, (P2 + P3 = 0.17), ω0 = 0.06, ω1 = 0.36, ω2 = 4.47 | 42 56 59 154138 142 163 165 4 15 134 147 158 176 |
| Branch leading to the SVP-like genes | 39 | 13.13** | P0 = 0.44, P1 = 0.37, (P2 + P3 = 0.19), ω0 = 0.06, ω1 = 0.37, ω2 = 2.01 | 16 14426170 2 4 7 55 84 129 132 133 184 186 188 197 |
| Type I | ||||
| Branch leading to PHERESI (AGL37) | 48 | 10.10** | P0 = 0.35, P1 = 0.41, (P2 + P3 = 0.24), ω0 = 0.10, ω1 = 0.32, ω2 = 6.52 | 105 92 7212057 99 63 118 78 88 66 89 47 |
Each comparison has n sequences. Proportions of the component site classes 0 (P0), 1 (P1), and 2 + 3 (P2 + P3), as well as the values for the background ratios ω0 and ω1 and the foreground ratio ω2, are given under MB. **, P < 0.005; ***, P < 0.001; bold underlined, PP ≥ 0.99 of being under positive selection; bold, 0.99 > PP ≥ 0.95; italics, 0.95 > PP ≥ 0.90; underlined, 0.90 > PP ≥ 0.70; normal, 0.70 > PP ≥ 0.50.
