Figure 3.
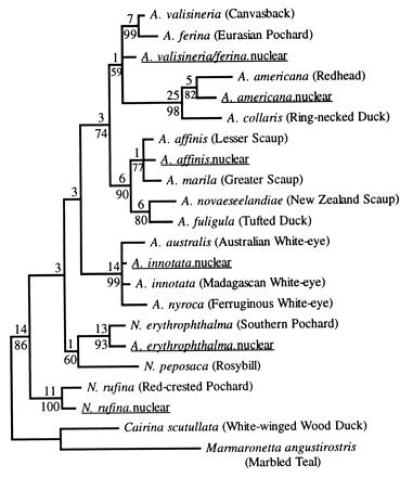
Strict consensus of 13 shortest trees (538 steps) found in a weighted parsimony analysis. The same set of shortest trees was found in all 100 random addition sequence replicates. Decay indices and bootstrap values are as in Fig. 2. Nuclear sequences homologous to the mtDNA control region are underlined. Cairina scutullata and Marmaronetta angustirostris were designated as the outgroup. We specified a step matrix that gave transitions, transversions, and indels weights of 1, 5, and 5, respectively. Note that branch lengths and decay indices reflect the weighted step matrix. Alignment gaps were introduced at 41 of 398 positions, but we excluded positions 127–133, 171–208, and 226–231 due to ambiguities in alignment, and positions 1–15 due to missing data.
