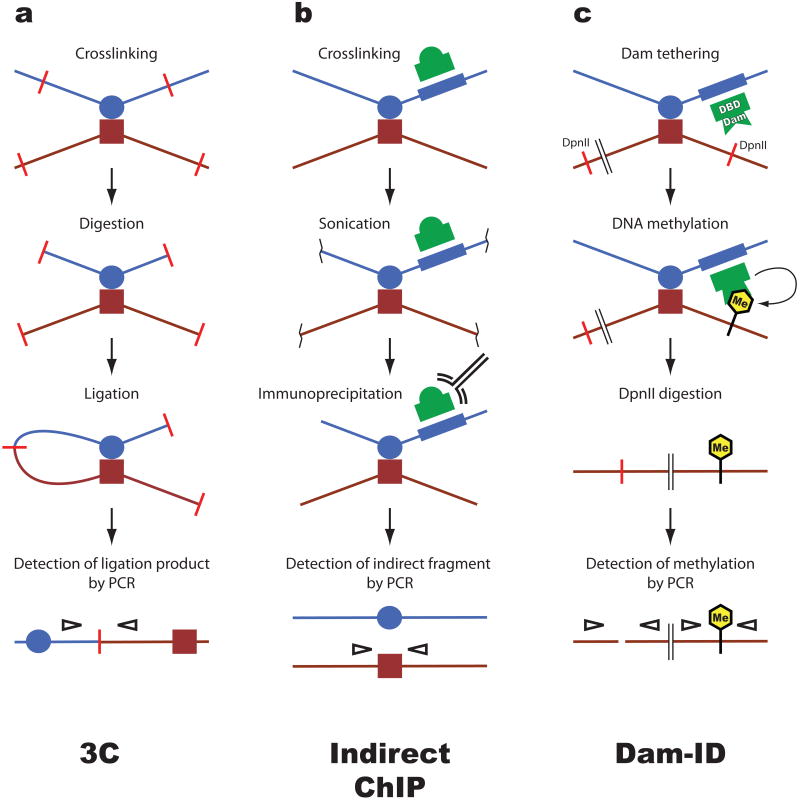
Figure 1. Methods to detect chromatin loops.
Several assays have been developed to detect proximity between two distant genomic sequences, for example an enhancer (blue circle) and a promoter (red square). a. Chromosome conformation capture (3C). This widely used assay relies on the assumption that sequences which interact with a high frequency can be crosslinked more efficiently than sequences that interact less frequently. Crosslinked chromatin is digested with a restriction enzyme that creates sticky ends (red bars). Ligation generates unique products which do not occur in genomic DNA and which can be detected by PCR (empty arrowheads). The amount of PCR product correlates with the proximity between the genomic fragments in vivo. b. Indirect chromatin immunoprecipitation (ChIP). This technique exploits sequence-specific DNA-binding proteins (green dome) as anchors to recover indirectly associated sequences. Chromatin is crosslinked and fragmented by sonication. DNA associated with the protein is recovered by immunoprecipitation. Sequences that are indirectly associated with the cognate sequence of the DNA-binding protein are recovered in this process and can be detected by PCR. c. DNA adenine methyltransferase identification (Dam ID). This method takes advantage of the fact that adenine methylation, which does not naturally occur in eukaryotes, inhibits DNA cleavage by the DpnII restriction enzyme. The bacterial Dam methylase is fused to a DNA-binding domain (DBD) to tether it to a sequence of interest (blue box). Tethered Dam methylates DNA in vivo, after which genomic DNA is digested with DpnII. PCR is used to amplify sequences in a fashion that distinguishes undigested i.e. methylated from unmethylated sites of interest.